-
MAPK Pathway
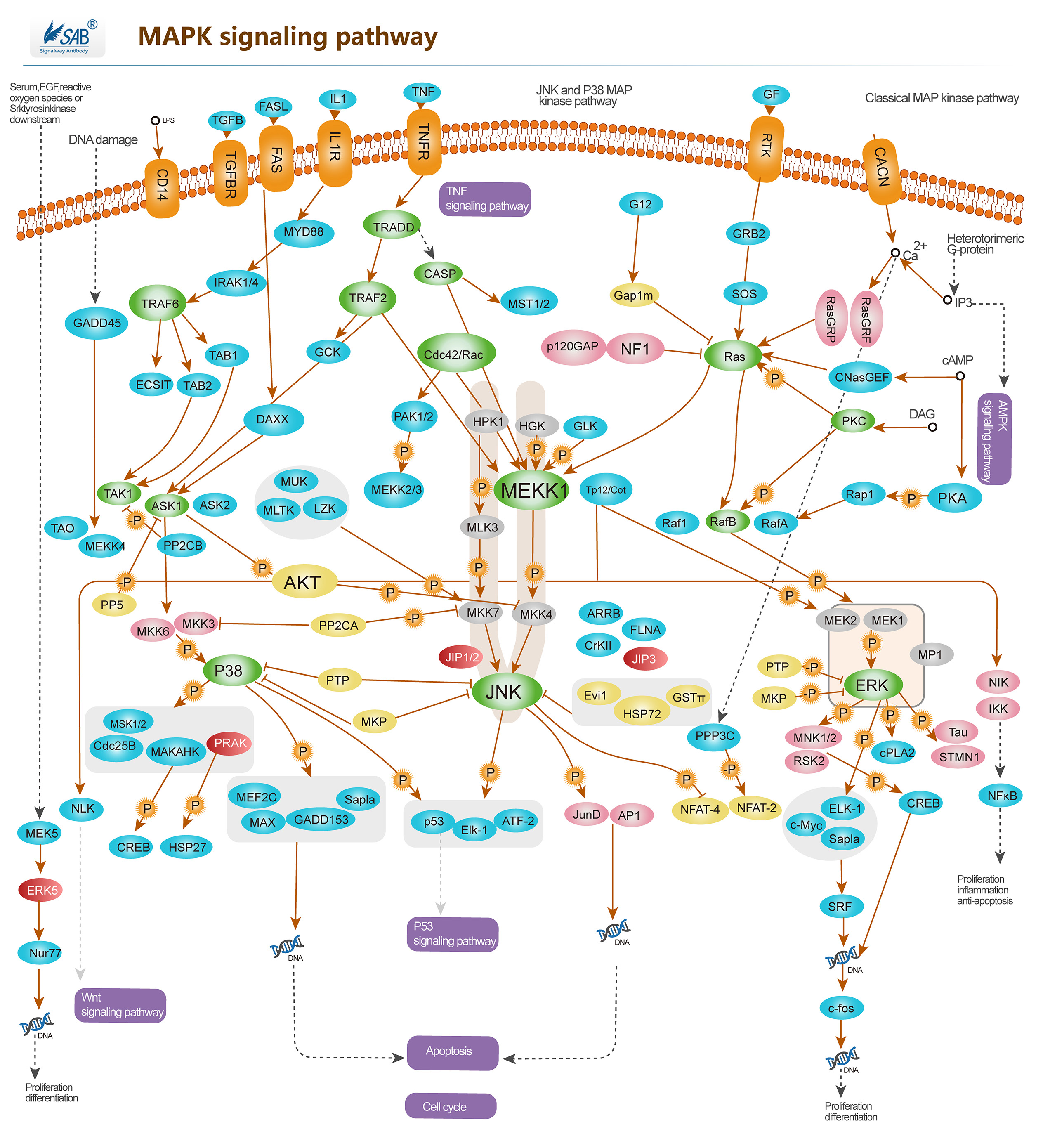 Pathway description:Intracellular signaling cascades are the main routes of communication between the Plasma membrane and regulatory targets in various intracellular compartments. Sequential activation of Kinases is a common mechanism of signal transduction in many cellular processes. During the past decade, several related intracellular signaling cascades have been elucidated, which are collectively known as MAPK (Mitogen-Activated Protein Kinase) signaling cascades. The MAPKs are a group of protein Serine/threonine Kinases that are activated in response to a variety of extracellular stimuli and mediate signal transduction from the cell surface to the nucleus. In combination with several other signaling pathways, they can differentially alter phosphorylation status of numerous proteins. There are four major groups of MAPKs in mammalian cells-the ERKs, the p38MAPKs, the JNKs and the ERK5 or BMK cascades. These MAPKs are actived by dual phosphorylation at the tripeptide motif Thr-Xaa-Tyr. Each MAPK pathway contains a three-tiered kinase cascade comprising a MAPKKK, a MAPKK and the MAPK. Frequently, a MAPKKKK activates the MAPKKK.The MAPKKK then phosphorylates a dual-specificity protein kinase MAPKK, which in turn phosphorylates the MAPK.p38MAPKs are members of the MAPK family that are activated by a variety of environmental stresses and inflammatory cytokines. Stress signals are delivered to this cascade by members of small GTPases of the Rho family (Rac, Rho, Cdc42). As with other MAPK cascades, the membrane-proximal component is a MAPKKK, typically a MEKK or a mixed lineage kinase (MLK). The MAPKKK phosphorylates and activates MKK3/6, the p38 MAPK kinase. MKK3/6 can also be activated directly by ASK1, which is stimulated by apoptotic stimuli. p38 MAPK is involved in regulation of Hsp27 and MAPKAP-2 and several transcription factors including ATF2, STAT1, the Max/Myc complex, MEF-2, ELK-1 and indirectly CREB via activation of MSK1.
Pathway description:Intracellular signaling cascades are the main routes of communication between the Plasma membrane and regulatory targets in various intracellular compartments. Sequential activation of Kinases is a common mechanism of signal transduction in many cellular processes. During the past decade, several related intracellular signaling cascades have been elucidated, which are collectively known as MAPK (Mitogen-Activated Protein Kinase) signaling cascades. The MAPKs are a group of protein Serine/threonine Kinases that are activated in response to a variety of extracellular stimuli and mediate signal transduction from the cell surface to the nucleus. In combination with several other signaling pathways, they can differentially alter phosphorylation status of numerous proteins. There are four major groups of MAPKs in mammalian cells-the ERKs, the p38MAPKs, the JNKs and the ERK5 or BMK cascades. These MAPKs are actived by dual phosphorylation at the tripeptide motif Thr-Xaa-Tyr. Each MAPK pathway contains a three-tiered kinase cascade comprising a MAPKKK, a MAPKK and the MAPK. Frequently, a MAPKKKK activates the MAPKKK.The MAPKKK then phosphorylates a dual-specificity protein kinase MAPKK, which in turn phosphorylates the MAPK.p38MAPKs are members of the MAPK family that are activated by a variety of environmental stresses and inflammatory cytokines. Stress signals are delivered to this cascade by members of small GTPases of the Rho family (Rac, Rho, Cdc42). As with other MAPK cascades, the membrane-proximal component is a MAPKKK, typically a MEKK or a mixed lineage kinase (MLK). The MAPKKK phosphorylates and activates MKK3/6, the p38 MAPK kinase. MKK3/6 can also be activated directly by ASK1, which is stimulated by apoptotic stimuli. p38 MAPK is involved in regulation of Hsp27 and MAPKAP-2 and several transcription factors including ATF2, STAT1, the Max/Myc complex, MEF-2, ELK-1 and indirectly CREB via activation of MSK1.
Selected Reviews:Wada T,Penninger JM.(2004)Mitogen-activated protein kinase in apoptosis regulation. Oncogene. 23(16),2838Panka DJ,Atkins MB,et al.(2006)Targeting the mitogen-activated protein kinase pathway in the treatment of malignant melanoma.Clin Cancer Res.12(7Pt2),2371s.Dunn KL, Espino PS, Drobic B, et al.(2005)The Ras-MAPK signal transduction pathway, cancer and chromatin remodeling. Biochem Cell Biol. 83(1):1-14 -
Akt Pathway
.jpg)
Pathway description:
Akt (v-Akt Murine Thymoma Viral Oncogene)/ PKB (Protein Kinase-B) is a Serine/threonine Kinase that is involved in mediating various biological responses, by phospho-rylation of a number of intracellular proteins, regulates different cellular processes, such as cell growth, cell cycle, apoptosis and glucose metabolism. Akt is activated by several hormones including insulin, growth factors, by signals derived from receptors for extracellular matrix molecules such as integrins, by several forms of cellular stress such as oxidative stress or cell swelling, and by activation of Ras. Three mammalian isoforms are currently known: Akt1/PKB- Alpha, Akt2/PKB-Beta and Akt3/ PKB-Gamma. All three isoforms of Akt share a common structure of three domains. Activation of PI3K by receptor tyrosine kinases mediates the phosphorylation of PIP2 to PIP3 and thereby the recruitment of Akt/PKB and PDK to the plasma membrane. Akt/PKB is subsequently activated and phosphorylation at Thr308 by PDK. PIP3 at the cell membrane recruits protein kinases such as Akt/PKB and PDK.PTEN functions as an antagonist of PI3K.PDK further activates SGK and aPKC , aPKC and SGK in turn phosphorylate a wide variety of cellular signaling molecules relevant for the regulation of cell growth, cell cycle and cell proliferation, including FKHR, GSK3, mTOR and p70S6K, for apoptosis, including Bad, caspase 9, IκB, FKHR, Mdm2. Akt/PKB further activates mTOR, a kinase stimulating the uptake of nutrients such as glucose, amino acids, cholesterol and iron. mTOR regulates the phosphorylation of p70S6K which can similarly be activated by PDK. mTOR further activates eIF4E-binding protein-1 and thus is involved in the regulation of translation.
Selected Reviews:Wang Q,Liu L,et al.(2003)Control of synaptic strength,a novel function of Akt.Neuron.38(6),915.Basso Ade, Solit DB,et al.(2002)Akt forms an intracellular complex with heat shock protein 90 (Hsp90) and Cdc37 and is destabilized by inhibitors of Hsp90 function.J Biol Chem.277(2),39858.Fornaro M,Plescia J,et al.(2003)Fibronectin protects prostate cancer cells from tumor necrosis factor-alpha-induced apoptosis via the AKT/survivin pathway. J Biol Chem.278(50),50402.Pommery N,Henichart JP.(2005)Involvement of PI3K/Akt pathway in prostate cancer –potential strategies for developing targeted therapies.Mini Rev Med Chem.5(12),1125 -
p53 signaling pathway
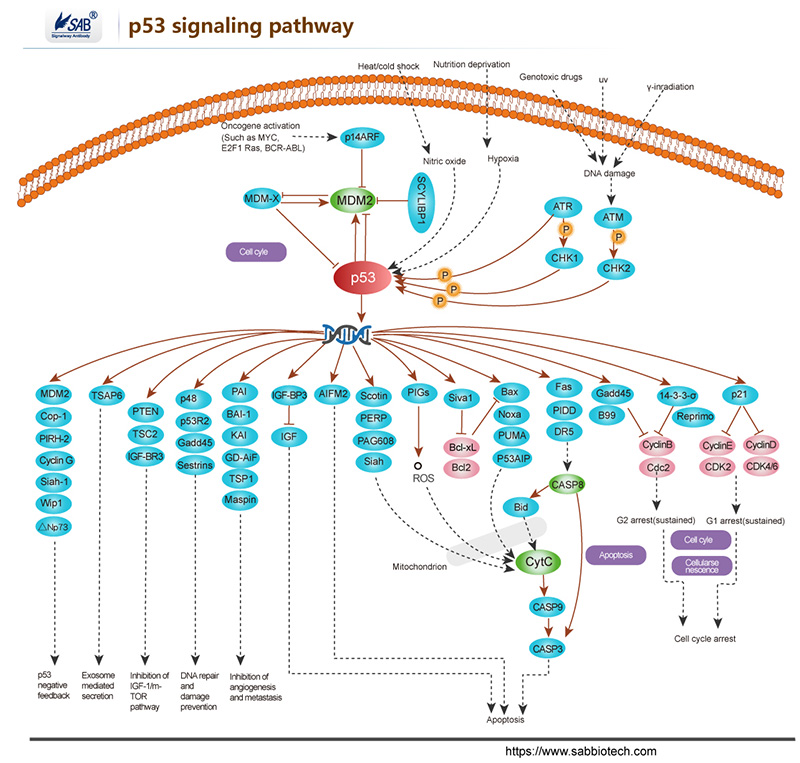
-
Hippo signaling pathway
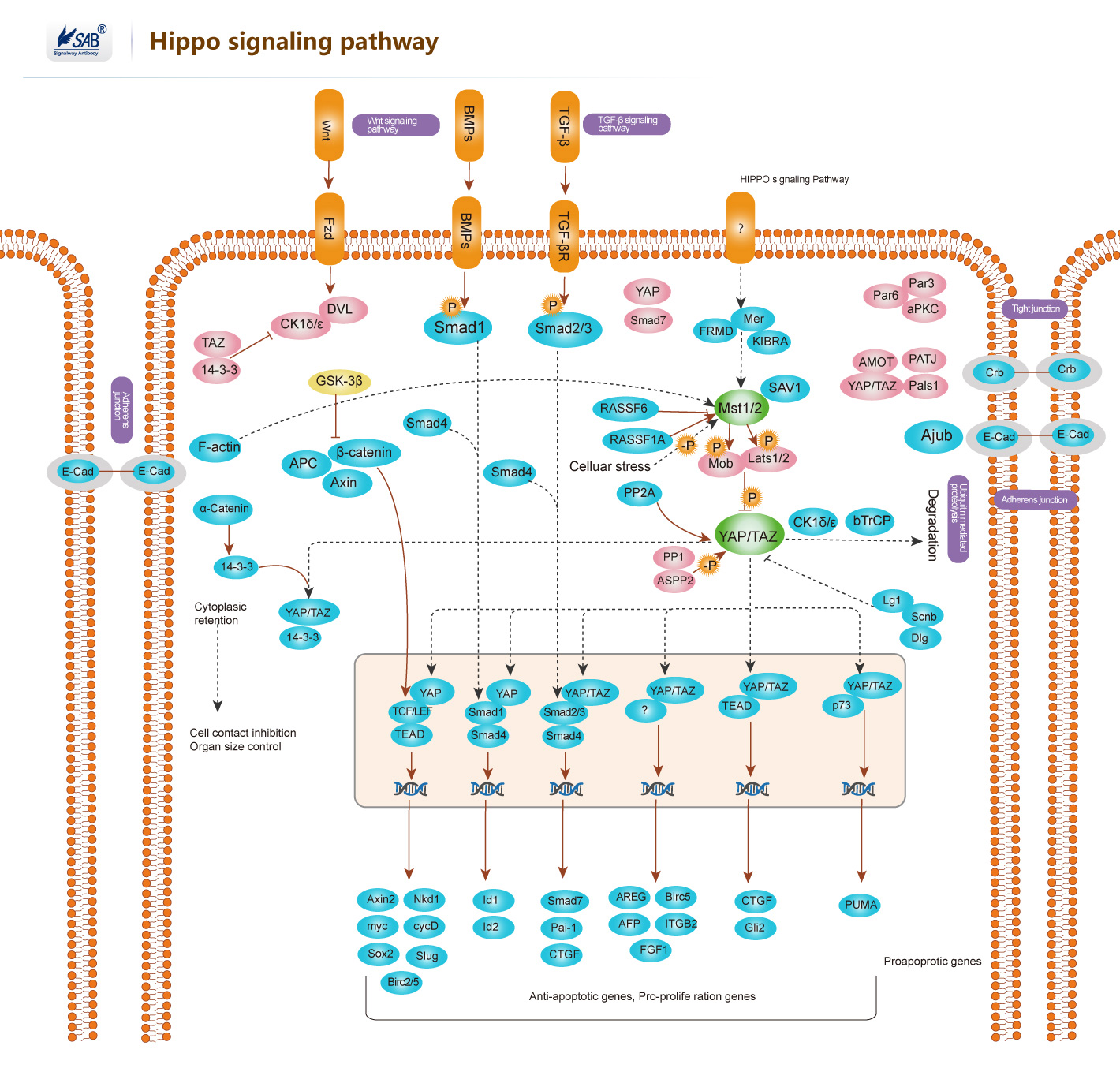
-
Wnt signaling pathway
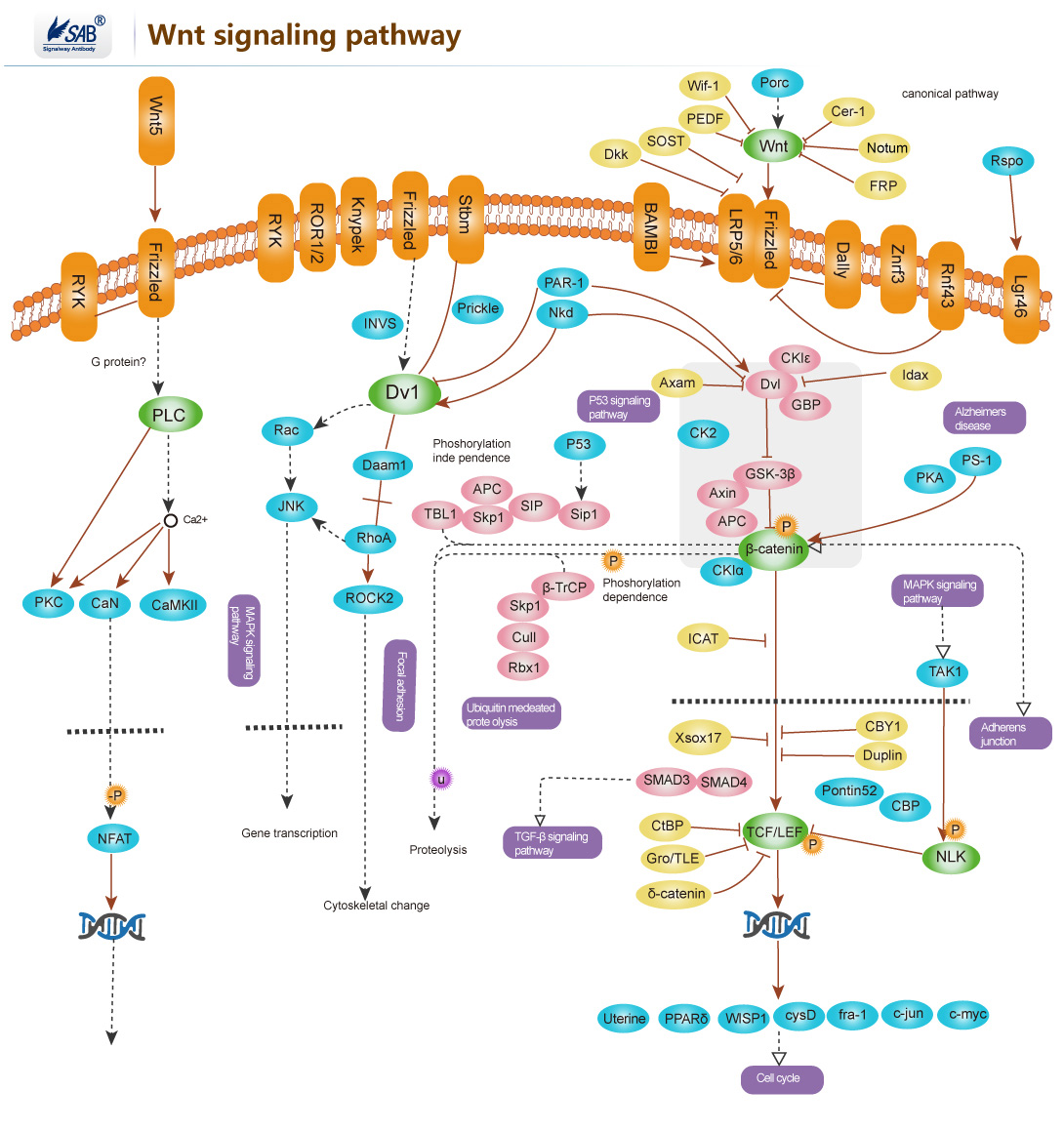
-
ErbB signaling pathway
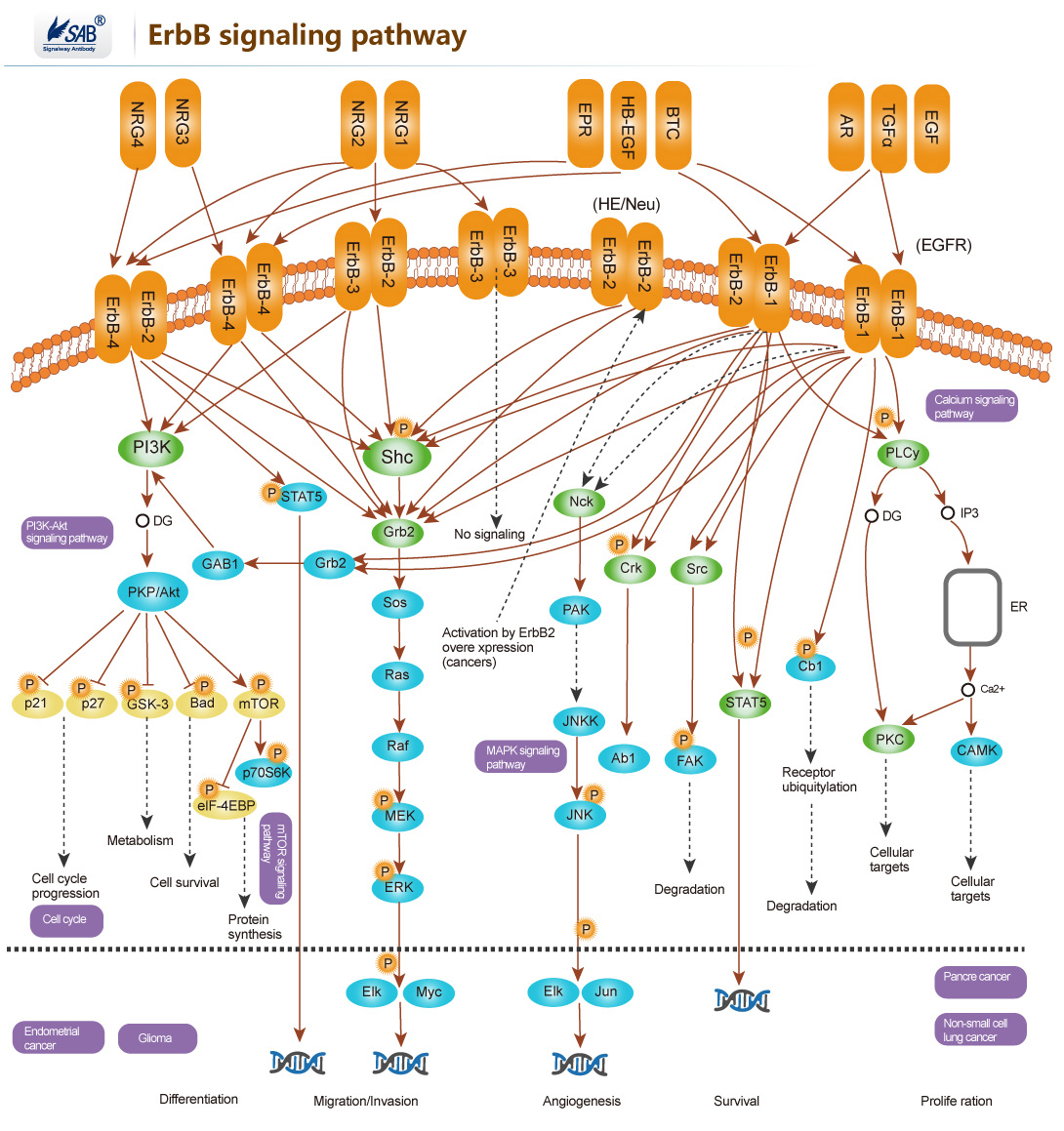
-
T-CELL Pathway
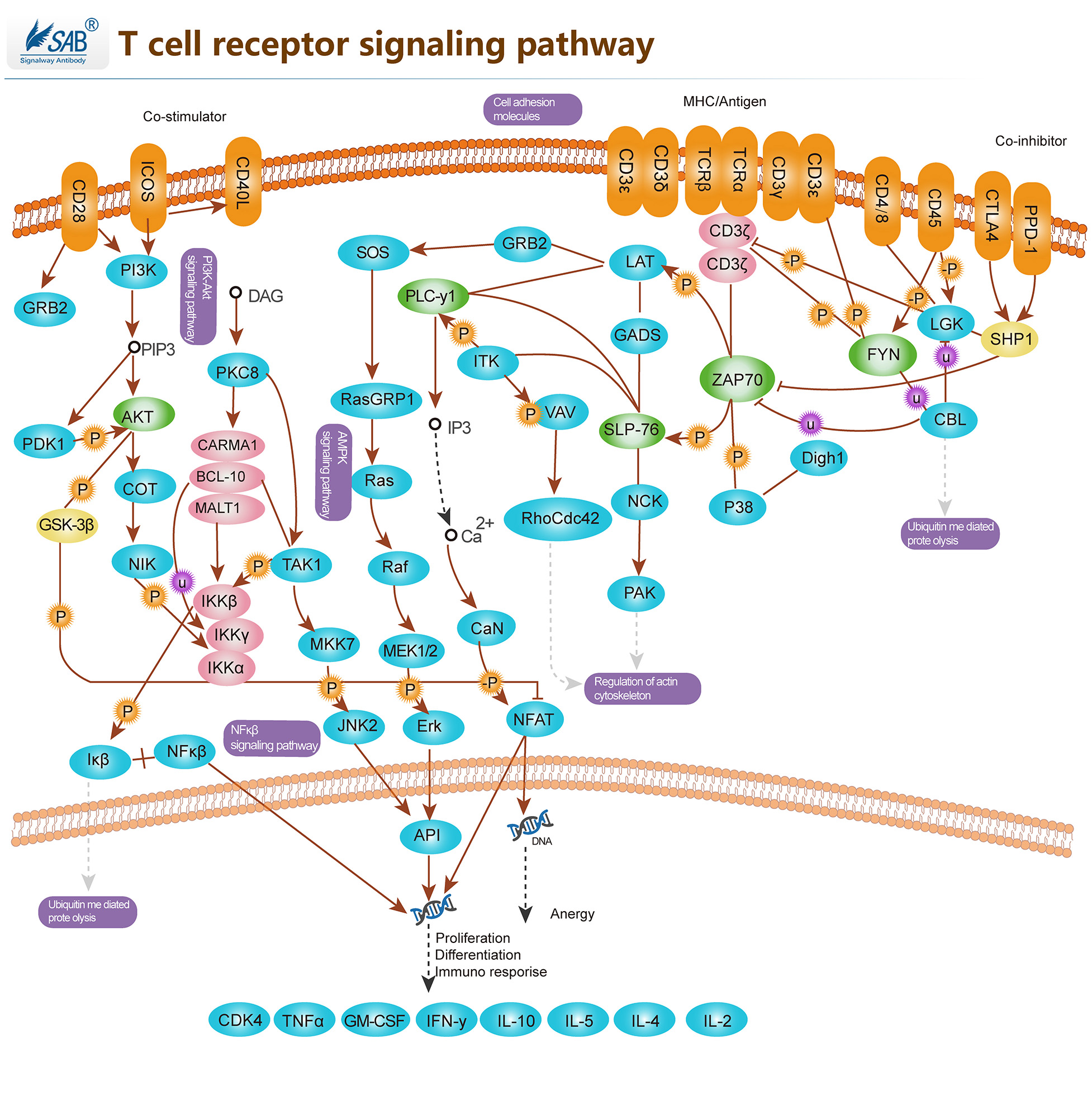
-
mTOR Pathway
.jpg)
-
JAK-STAT Pathway
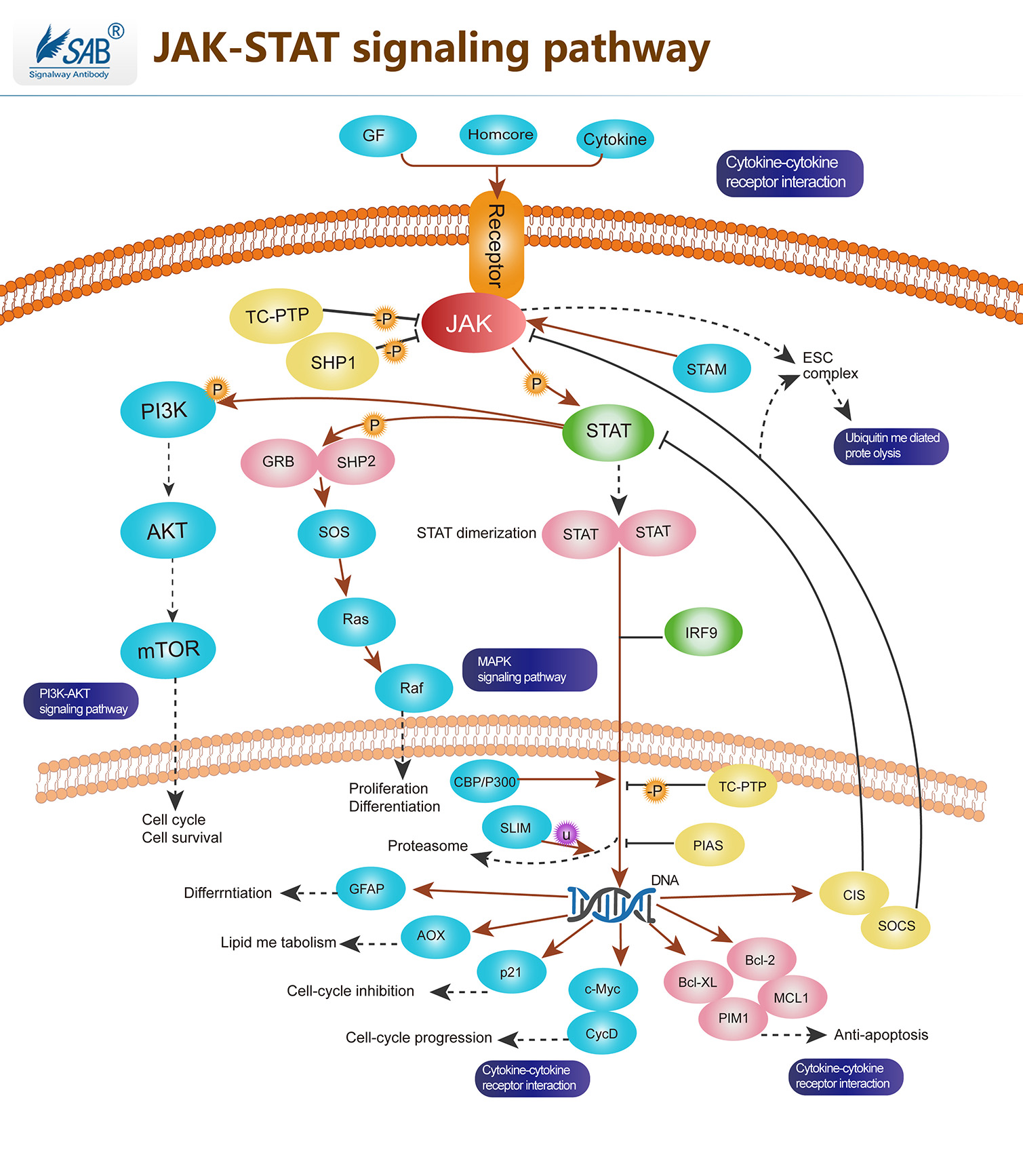 Pathway description:The Janus Kinase (JAK) and signal transducers and activators of transcription (STAT) pathway has been shown to play a key role in cytokine-mediated signal transduction, and to regulate growth, differentiation, and death of both normal and transformed cells.The JAK/STAT pathway represents one such signaling cascade whose evolutionarily conserved roles include cell proliferation and haematopoiesis. Jak belongs to a family of non-receptor protein tyrosine kinase of approximately 130kDa, comprising of Jak1, Jak2, Jak3 and Tyk2.JAK activation occurs upon ligand-mediated receptor multimerization because two JAKs are brought into close proximity, allowing trans-phosphorylation. The activated JAKs subsequently phosphorylate additional targets, including both the receptors and the major substrates, STATs. STATs are latent transcription factors that reside in the cytoplasm until activated. The seven mammalian STATs bear a conserved tyrosine residue near the C-terminus that is phosphorylated by JAKs. This phosphotyrosine permits the dimerization of STATs through interaction with a conserved SH2 domain. Phosphorylated STATs enter the nucleus by a mechanism that is dependent on importin -5 (also called nucleoprotein interactor 1) and the Ran nuclear import pathway.Once in the nucleus, STAT dimers bind specific enhancers, regulating the transcription of target genes. In addition to activating STATs, Jak kinases phosphorylate other signaling/adaptor proteins, linking Jak signaling to other pathways such as the MAP kinases. These parallel regulatory pathways are important in shaping the specificity of cellular responses to various cytokines.Selected Reviews:Ananthakrishnan R,Hallam K,et al.(2005)JAK-STAT pathway in cardiac ischemic stress.Vascul Pharmacol.Walker JG,Smith MD.(2005)The JAK-STAT pathway in rheumatoid arthritis.J Rheumatol.32(9),1650-3.Rawlings JS, Rosler KM, Harrison DA.(2004)The JAK/STAT signaling pathway. J Cell Sci. 117(Pt 8):1281-3.
Pathway description:The Janus Kinase (JAK) and signal transducers and activators of transcription (STAT) pathway has been shown to play a key role in cytokine-mediated signal transduction, and to regulate growth, differentiation, and death of both normal and transformed cells.The JAK/STAT pathway represents one such signaling cascade whose evolutionarily conserved roles include cell proliferation and haematopoiesis. Jak belongs to a family of non-receptor protein tyrosine kinase of approximately 130kDa, comprising of Jak1, Jak2, Jak3 and Tyk2.JAK activation occurs upon ligand-mediated receptor multimerization because two JAKs are brought into close proximity, allowing trans-phosphorylation. The activated JAKs subsequently phosphorylate additional targets, including both the receptors and the major substrates, STATs. STATs are latent transcription factors that reside in the cytoplasm until activated. The seven mammalian STATs bear a conserved tyrosine residue near the C-terminus that is phosphorylated by JAKs. This phosphotyrosine permits the dimerization of STATs through interaction with a conserved SH2 domain. Phosphorylated STATs enter the nucleus by a mechanism that is dependent on importin -5 (also called nucleoprotein interactor 1) and the Ran nuclear import pathway.Once in the nucleus, STAT dimers bind specific enhancers, regulating the transcription of target genes. In addition to activating STATs, Jak kinases phosphorylate other signaling/adaptor proteins, linking Jak signaling to other pathways such as the MAP kinases. These parallel regulatory pathways are important in shaping the specificity of cellular responses to various cytokines.Selected Reviews:Ananthakrishnan R,Hallam K,et al.(2005)JAK-STAT pathway in cardiac ischemic stress.Vascul Pharmacol.Walker JG,Smith MD.(2005)The JAK-STAT pathway in rheumatoid arthritis.J Rheumatol.32(9),1650-3.Rawlings JS, Rosler KM, Harrison DA.(2004)The JAK/STAT signaling pathway. J Cell Sci. 117(Pt 8):1281-3. -
B CELL Pathway
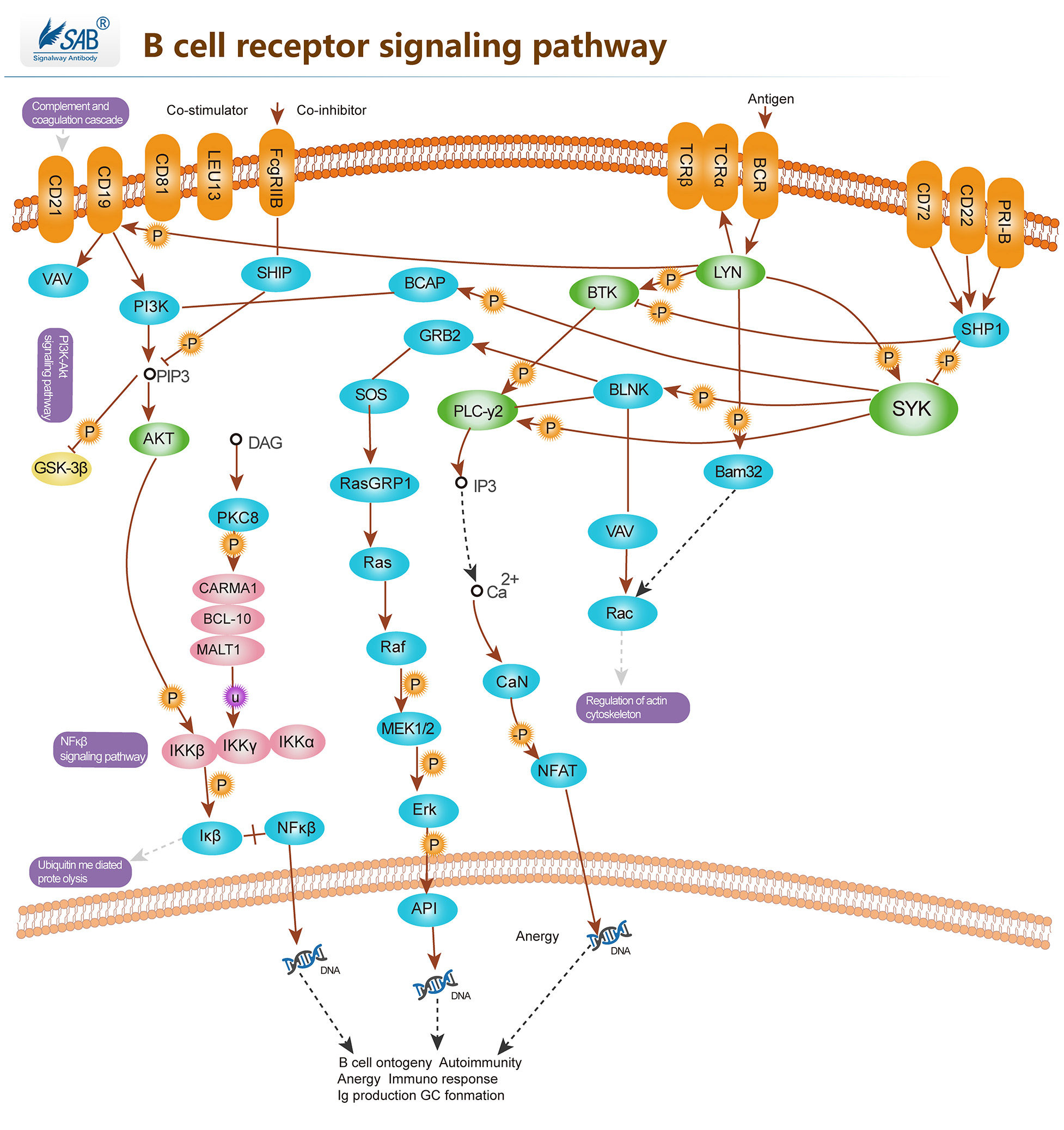
-
Autophagy Pathway
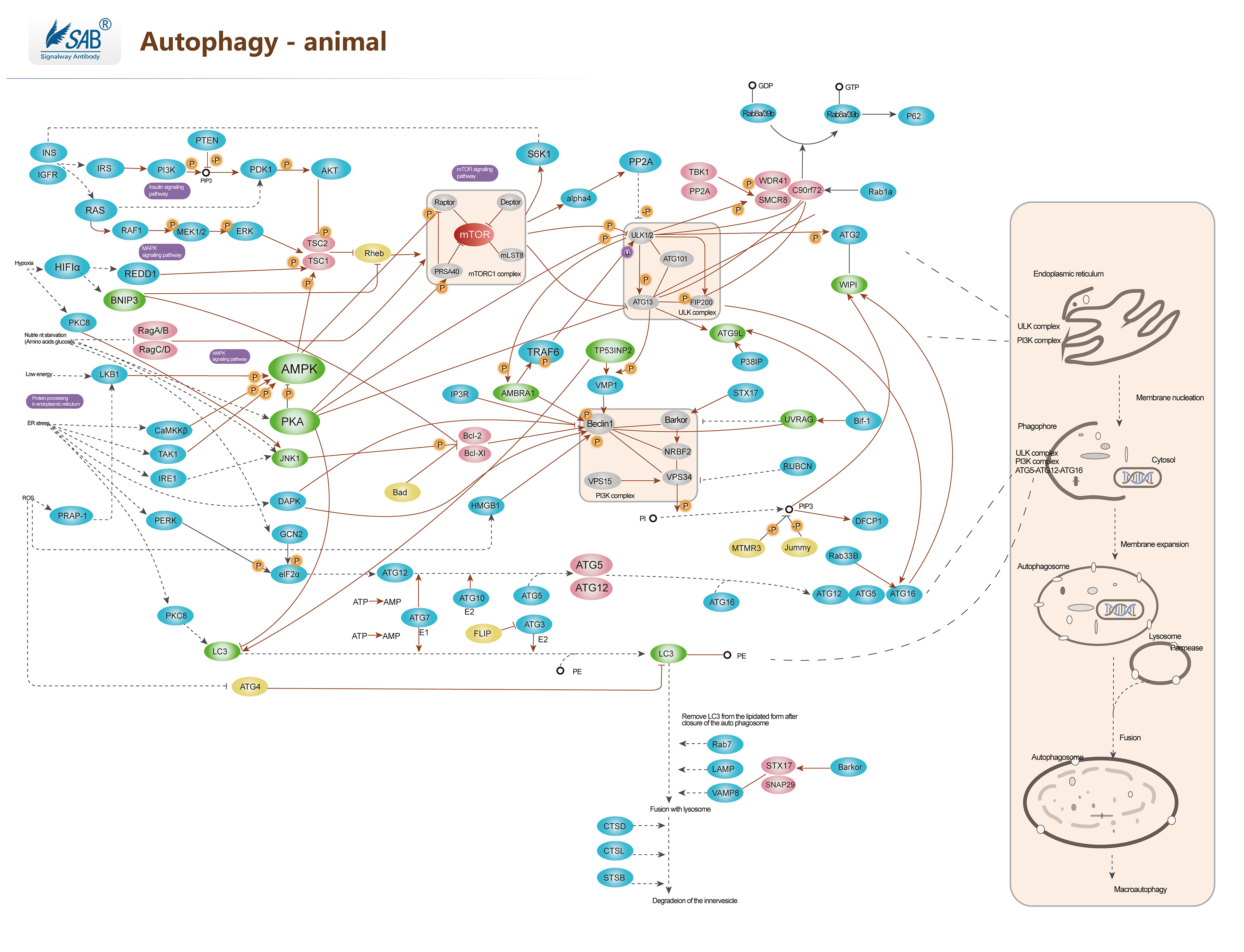
-
AMPK Pathway
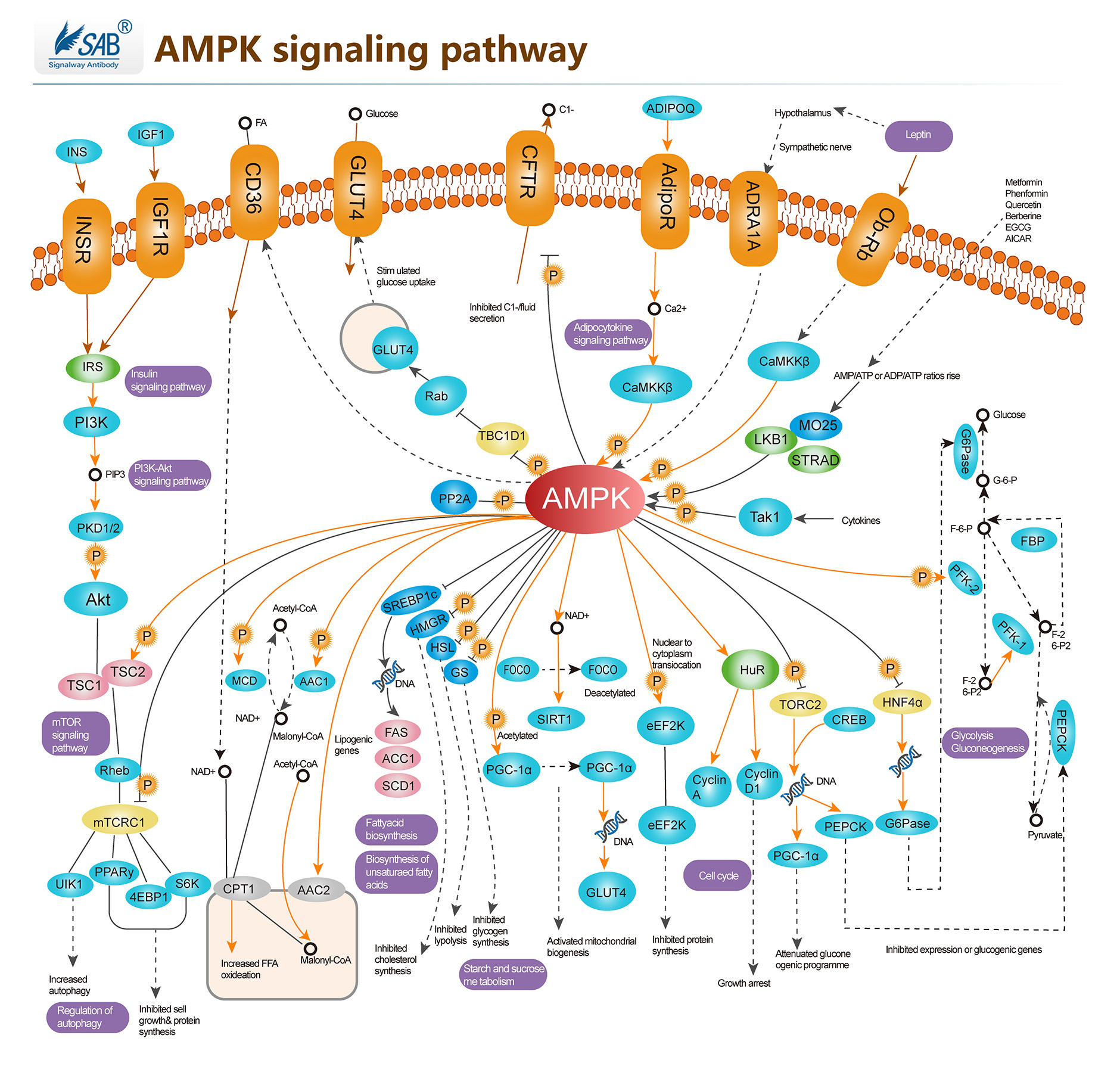
-
Troubleshooting tips for IHC
Troubleshooting tips for IHC common problems:
1. Non-specifc staining
2. No staining
3. Weak staining
4. Strong Staining
1.Non-specifc staining
Causes Solutions Improper preparation of sections Improve ways of sampling and preparation Inadequate deparaffinization of the sections Increase the deparaffinization time Tissue contains endogenous peroxidase Use 0.3% v/v fresh H2O2 for blocking and increase the blocking incubation time Tissue contains endogenous biotin Use IHC biotin blocking agent Blocking of protein may be insufficient Increase the blocking time Charge adsorption Block with nonimmune animal serum The antibody is not pure Change suitable antibody Primary antibody concentration may be too high Try decreasing the antibody concentration The sections have dried out Avoid sections being dried out in the process of experiments Washes may be insufficient Increase the times of washes and the washing time Non-specifc staining: Improved: 
Immunohistochemical analysis of paraffin-embedded human lung carcinoma tissue showing cytoplasmic and nuclear staining using NFκB-p65 Phospho-Ser276 Antibody #11011.
2.No staining
Causes Solutions Improper tissue processing Try to improve the condition and sampling again No antigen in the tissue Set a positive control to verify the experiment results The antibody is not active Don’t use out-of-date antibody kits Incompatible secondary and primary antibodies Use secondary antibody that was raised against the species in which the primary was raised Incompatible staining system Change compatible staining system Improper operation and leave out important steps Follow strict operating procedure and set a positive control No staining: Improved: 

Immunohistochemical analysis of paraffin-embedded human breast carcinoma tissue using Histone H3 Di-Methyl-Lys27 Antibody #11583.
3.Weak staining
Causes Solutions Improper tissue fixation or too high temperature when fixing Use appropriate fixation way or fixation time Too high baking slides temperature and too long baking time Choose appropriate temperature and time for baking slides The antigen may be damaged Let fresh tissues be fixed in time and for not to exceed 24 hours Over blocking of protein Reduce the blocking time The antibody has drained away Ensure that the sections are placed in a horizontal position when incubating The antibody concentration may be too low or incubation time may be too short Increase the antibody concentration and incubation time The room temperature may be too low. Lower than 15℃. Incubator at 37℃ or increase incubation time No draining off buffer solution when adding the reagent results in the reagent being diluted. Do drain off buffer solution but avoid sections being dried out. Excessive washing Wash moderately Always verify the expiration date of the reagent prior to use Change reagents timely Weak staining: Improved: 
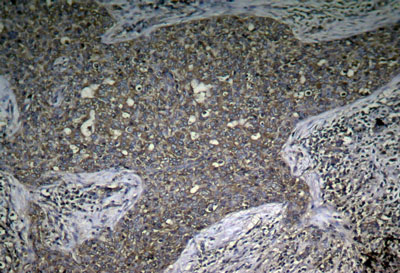
Immunohistochemical analysis of paraffin-embedded human breast carcinoma tissue using mTOR Phospho-Ser2448 Antibody#11221.
4.Strong Staining
Causes Solutions The primary antibody concentration may be too high or incubation time may be too long Reduce the primary antibody concentration or incubation time. The incubation temperature may be too high Incubate at 4℃ or at room temperature The incubation time of HRP conjugated secondary antibody may be too long Reduce the incubation time Inadequate washing Increase the times of washing Strong Staining: Improved: .jpg)
.jpg)
Immunohistochemical analysis of paraffin-embedded human breast carcinoma tissue using FKHR Phospho-Ser256 Antibody#11115.
-
Troubleshooting tips for western blotting
Troubleshooting tips for western blotting common problems:1.High background2.Low or no signal3.Non-specific bands4.Wrong band location5.Invisible dots on the bands6.Incomplete bands7.Smile effect of the bands8.White bands on a black blot9.Black dots on the blot10.Marker lane is black1.High Background
Causes Solutions Membrane fouling Use clean tweezer and operate with gloves to prevent membrane fouling. The membrane has dried out Incubate in sufficient reaction solution to prevent the membrane from drying out. Blocking insufficient Increase the blocking incubation time Inappropriate blocking buffer Switch different blocking buffer Buffer solution has been contaminated Use new buffer solution Incomplete washing Increase washing time and washing buffer’s volume The antibody concentration may be too high Decrease the concentration of primary antibody or secondary antibody Cross-reaction between antibody and blocking agent Choose blocking solution without cross-reaction. Add Tween-20 to the washing buffer to reduce cross reaction. High background: 
Improved: .gif)
2. Low or no signal
Causes solutions Insufficient antigen Increase amount of loading samples Protein degradation Re-prepare samples The antigen is blocked by blocking buffer Optimize blocking solution, decrease blocking time or decrease the concentration of proteins in the blocking solution. No or low level of target protein in samples Run a positive control. If the level of target protein in samples is low, try to increase amount of loading sample. Wrong choice of membrane Choose suitable pore size membrane. Use 0.45um size membrane for proteins larger than 22KD. Use 0.2 µm size membrane for proteins smaller than 22 KD. Poor transfer of protein to membrane Make sure there are no air bubbles between the gel and membrane during transfer. Always ensure assembling electrode correctly. Control transfer temperature and optimize transfer electricity and time. Methanol concentration may be too high. Too high concentration of methanol may result in the separation of protein and SDS and thus cause protein precipitation in the gel. At the same time, it may cause shrinking or hardening of the gel to inhibit transferring of high molecular weight proteins. As a result, choose suitable methanol concentration according to different molecular weight. Excessive washing of membrane Reduce washing time and washing times Over blocking Lower the concentration of your blocking solution and shorten blocking time. Change blocking solutions. The primary antibody is inactive Use effective antibody in expiration, avoid freezing- thawing repeatedly, and use fresh solution. Insufficient reaction of antibody to membrane Increase the concentration of the antibody and the incubation time. The reagents are not compatible with each other Primary antibody and species, primary antibody and secondary antibody, or enzyme and substrate are not compatible. Setting loading control can validate the secondary detecting system. HRP Inhibited Avoid sodium azide in all solutions and containers Enzyme or substrate is inactive. Directly mix enzyme and substrate. If no color, the enzyme doesn’t work. Choose active conjugated reagent. Use fresh substrate. Exposure time is too short Increase the exposure time Low or no signal Improved 
3. Non-specific bands
Causes Solutions Amount of loading samples is too large Decrease amount of loading samples Protein degradation Use fresh samples and use protein inhibitor Cells were cultured too many passages to result in protein variation Use primary cells or less passaging cells to run a control Antibody Concentration is too high Decrease the concentration of the primary or secondary antibody Cross-reaction Choose monoclonal antibody or affinity purified antibody to ensure antibody specificity Existence of the different protein isoforms Some proteins derived from the same gene have different isoforms. The size of every isoform protein is different. Non-specific signal caused by the secondary antibody Run a secondary antibody control or choose other secondary antibody Substrate is too sensitive Use suitable substrate Exposure time is too long Reduce the exposure time Non-specific bands: Improved: 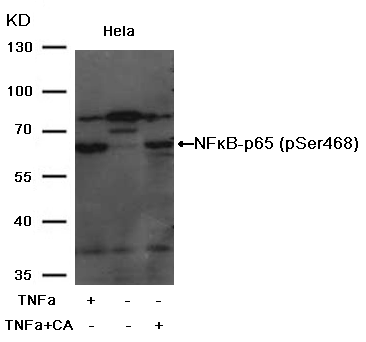
4.Wrong band location
Causes Solutions Existence of dimer or polymer Increase a process or intensity of protein denaturation Modification of proteins Modification of proteins, such as glycosylation or phosphorylation, can result in an increase of molecular weight of protein. 5.Invisible dots on the bands
Causes Solutions Air bubbles were trapped in the gap of gel and membrane during transferring. Remove bubbles in the gap of gel and membrane when preparing for transferring. Invisible dots on the bands: Improved: 
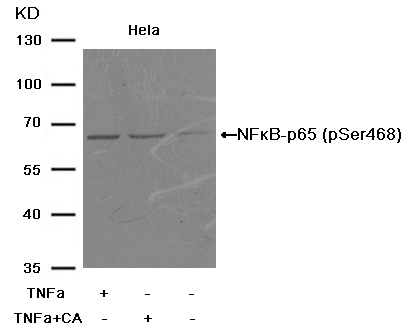
6.Incomplete bands
Causes Solutions Substrate is not well-distributed during incubation Even the substrate during incubation Incomplete bands: Improved: 

7.Smile effect of the bands
Causes Solutions Migration was too fast during electrophoresis Reduce the voltage to slow down the migration Migration was too hot Run the gel in the cold room or in ice Smile effect of the bands: Improved: 

8.White bands on a black blot
Causes Solutions HRP concentration is too high Decrease the secondary antibody concentration White bands on a black blot: Improved: 

9.Black dots on the blot
Causes Solutions Air bubbles were trapped against the membrane during transferring or the antibody is not well distributed during incubation Try to remove bubbles. Keep shaking when incubating the antibody. The blocking agent was not well dissolved. Filter the blocking agent. The antibody reacts with the blocking solution. Choose suitable blocking solution There are aggregates in the HRP conjugated secondary antibody. Filter the secondary antibody agent and remove the aggregates. Black dots on the blot: Improved: 
10.Marker lane is black
Causes Solutions The antibody reacts with the MW marker Add a blank lane between the marker and the adjacent sample lane -
Flow Cytometry(FC)
Flow Cytometry (FC) is a new technology for analysis on individual cells by means of flow cytometers. It allows quantitative analysis by measuring multiple parameters simultaneously. Characterized by without damage to the structure and function of cells, organelles or analytical particles in the measurement and speedy, accurate, sensitive and quantifiable process, it has been widely used in cell counting, cell sorting, and analysis of the molecules on cell surface or intracellular. SAB follows strict validation procedure on every batch of antibody to ensure accurate and credible results in FC.
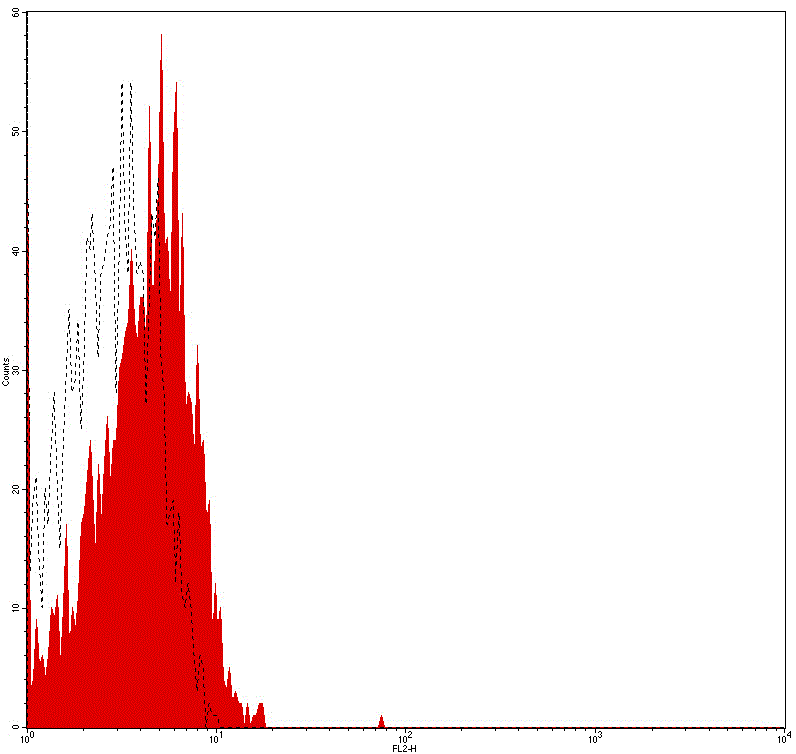
Flow cytometry analysis of Human peripheral blood lymphocytes using Mouse IgG2b Isotype Control, FITC Conjugated mAb #28279.
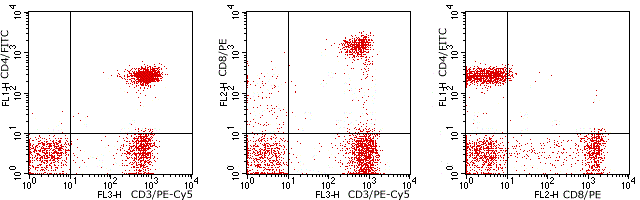
Flow cytometry analysis of Human peripheral blood lymphocytes using Human CD4/CD8/CD3, FITC/PE/PE-Cy5 Conjugated mAb #28249.
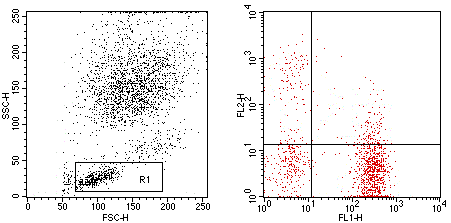
Flow cytometry analysis of Human peripheral blood lymphocytes using Human CD3/HLA-DR, FITC/PE Conjugated mAb #28239.

Flow cytometry analysis of Human peripheral blood monocytes using Human CD64, PE Conjugated mAb #28198.
-
Immunofluorescence (IF)
Immunofluorescence (IF) is used to analyze the sub-cellular localization of a protein within a cell. This technique usually uses a labeled antibody to target the fluorescent dye to its antigen. For the emitted light by the fluorescent dye can be detected in fluorescence microscope, this allows visualization of the distribution of a target protein in the sample. SAB follows stringent validation protocol to ensure IF application.
Immunofluorescence (IF) validation methods:Using appropriate cell lines for analysisStimulus treatmentInhibitor treatmentUsing appropriate cell lines for analysis Immunofluorescence analysis of methanol-fixed MCF7 cells using HER2 (Phospho-Tyr1248) Antibody #11079.
Immunofluorescence analysis of methanol-fixed MCF7 cells using HER2 (Phospho-Tyr1248) Antibody #11079.
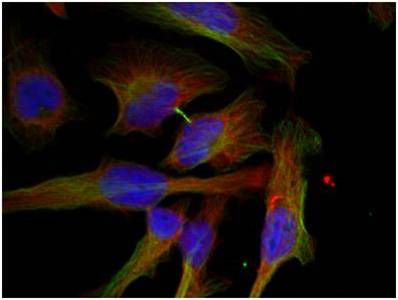 Immunofluorescence analysis of methanol-fixed Hela cells using GSK3β (Phospho-Ser9) Antibody #11002.
Immunofluorescence analysis of methanol-fixed Hela cells using GSK3β (Phospho-Ser9) Antibody #11002.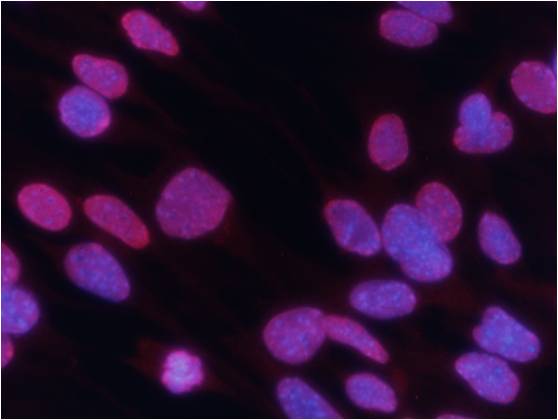 Immunofluorescence analysis of methanol-fixed MEF cells using Histone H3 (Tri-Methyl-Lys27) Antibody #11582.Stimulus treatment
Immunofluorescence analysis of methanol-fixed MEF cells using Histone H3 (Tri-Methyl-Lys27) Antibody #11582.Stimulus treatment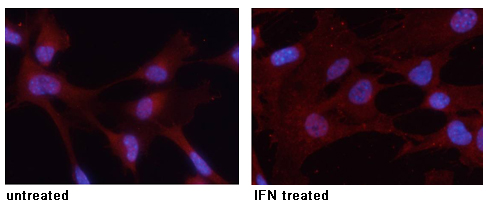 Immunofluorescence analysis of methanol-fixed MEF cells untreated or treated with IFN using STAT3 (Phospho-Tyr705) Antibody #11045.
Immunofluorescence analysis of methanol-fixed MEF cells untreated or treated with IFN using STAT3 (Phospho-Tyr705) Antibody #11045.Inhibitor treatment
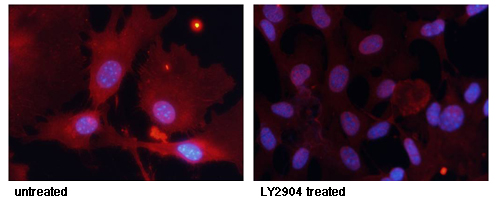
Immunofluorescence staining of methanol-fixed MEF cells untreated or treated with LY2904 using FKHR (Phospho-Ser256) Antibody #11115.
-
Immunohistochemistry(IHC)
Immunohistochemistry (IHC) is commonly used for in situ qualitative or quantitative analysis of a target protein in cells of a tissue section. Thought a labeled antibody that is specific to its antigen and sensitive to color-developing agent, this method is also widely used for analyzing the sub-cellular localization of a protein. SAB offers IHC applicable antibodies which have generally been validated by different tissues and various approaches. This will distinctly save your time in experiment and can avoid waste of samples.
Validation Approaches Include:Validation by different tissuesBlocking peptide controlHuman breast carcinoma tissue Immunohistochemical analysis of paraffin-embedded human breast carcinoma tissue using NFκB-p65 (Phospho-Ser536) Antibody #11014.Human Lung carcinoma tissue
Immunohistochemical analysis of paraffin-embedded human breast carcinoma tissue using NFκB-p65 (Phospho-Ser536) Antibody #11014.Human Lung carcinoma tissue Immunohistochemical analysis of paraffin-embedded human Lung carcinoma tissue using NFκB-p65 (Phospho-Ser536) Antibody #11014.
Immunohistochemical analysis of paraffin-embedded human Lung carcinoma tissue using NFκB-p65 (Phospho-Ser536) Antibody #11014.Human colon carcinoma tissue
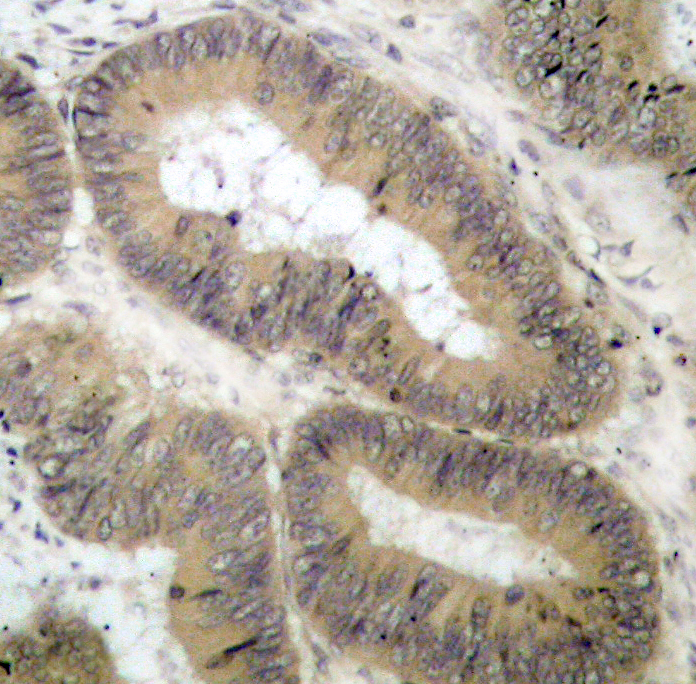 Immunohistochemical analysis of paraffin-embedded human colon carcinoma tissue using IKK α (Phospho-Thr23) Antibody #11129.Human tonsil carcinoma tissue
Immunohistochemical analysis of paraffin-embedded human colon carcinoma tissue using IKK α (Phospho-Thr23) Antibody #11129.Human tonsil carcinoma tissue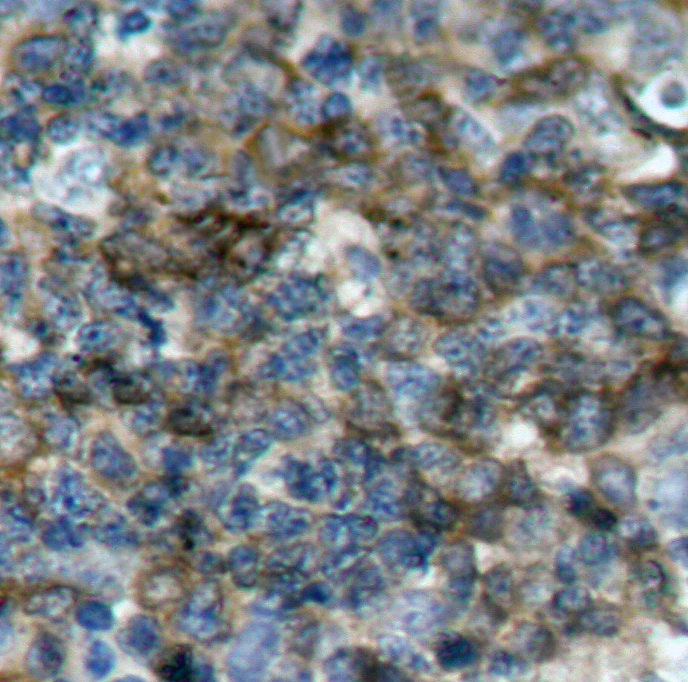 Immunohistochemical analysis of paraffin-embedded human tonsil carcinoma tissue using VASP (Phospho-Ser157) Antibody #11214Rat hippocampal region tissue
Immunohistochemical analysis of paraffin-embedded human tonsil carcinoma tissue using VASP (Phospho-Ser157) Antibody #11214Rat hippocampal region tissue Immunohistochemical analysis of paraffin-embedded rat hippocampal region tissue from a model with Alzheimer’s Disease using Tau (Phospho-Thr205) Antibody #11108.Blocking peptide control
Immunohistochemical analysis of paraffin-embedded rat hippocampal region tissue from a model with Alzheimer’s Disease using Tau (Phospho-Thr205) Antibody #11108.Blocking peptide control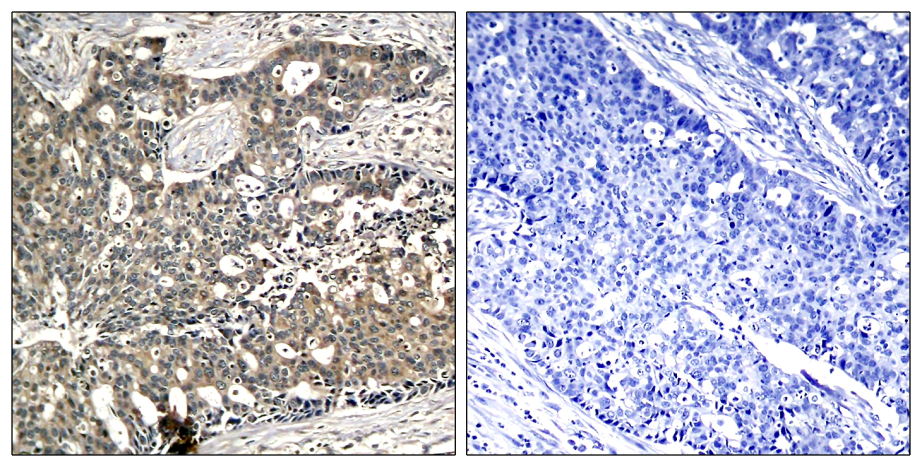 Immunohistochemical analysis of paraffin-embedded human breast carcinoma tissue using p56Dok-2 (Phospho-Tyr299) Antibody #11278 (left) or the same antibody preincubated with blocking peptide #51278 (right).
Immunohistochemical analysis of paraffin-embedded human breast carcinoma tissue using p56Dok-2 (Phospho-Tyr299) Antibody #11278 (left) or the same antibody preincubated with blocking peptide #51278 (right).
-
Western blotting(WB)
(1).gif)
Western blotting (WB) is used to analyze a target protein in a mixed sample with numerous proteins according to the specificity of antibody against its antigen. With SDS-PAGE’s high resolution and solid-phase immunoassay’s high sensitivity and specificity, western blotting has been one of the most common methods for protein analysis. SAB follows a stringent validation protocol and uses several validation approaches in western blotting to ensure antibody quality and phospho-specificity.
Validation Approaches Include:
Phosphatase treatment
Stimulus or inhibitor treatment
Blocking peptide control
Validation by various cell lines
Phosphatase treatment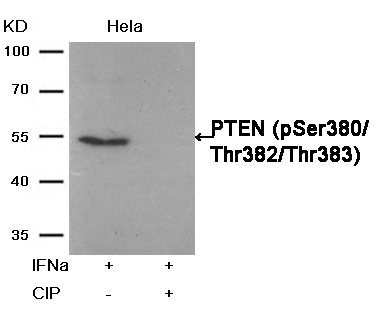 Western blot analysis of extracts from Hela cells, treated with IFNa or calf intestinal phosphatase (CIP), using PTEN (Phospho-Ser380/Thr382/Thr383) Antibody #11056.Stimulus or inhibitor treatment
Western blot analysis of extracts from Hela cells, treated with IFNa or calf intestinal phosphatase (CIP), using PTEN (Phospho-Ser380/Thr382/Thr383) Antibody #11056.Stimulus or inhibitor treatment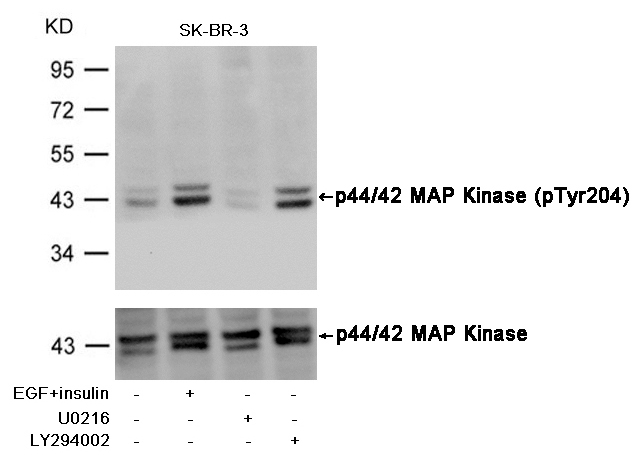 Western blot analysis of extracts from SK-BR-3 cells, untreated or insulin and EGF treated, and pretreated with U0126 and LY294002, using p44/42 MAP Kinase (Phospho-Tyr204) Antibody #11246 (upper) or p44/42 MAP Kinase Antibody #21238 (lower) .Fluorescent Western Blotting
Western blot analysis of extracts from SK-BR-3 cells, untreated or insulin and EGF treated, and pretreated with U0126 and LY294002, using p44/42 MAP Kinase (Phospho-Tyr204) Antibody #11246 (upper) or p44/42 MAP Kinase Antibody #21238 (lower) .Fluorescent Western Blotting Western blot analysis of extracts from 293 cells, treated with Hydroxyurea or calf intestinal phosphatase(CIP), using CDC2 (Phospho-Tyr15) Antibody . #11244.Blocking peptide control
Western blot analysis of extracts from 293 cells, treated with Hydroxyurea or calf intestinal phosphatase(CIP), using CDC2 (Phospho-Tyr15) Antibody . #11244.Blocking peptide control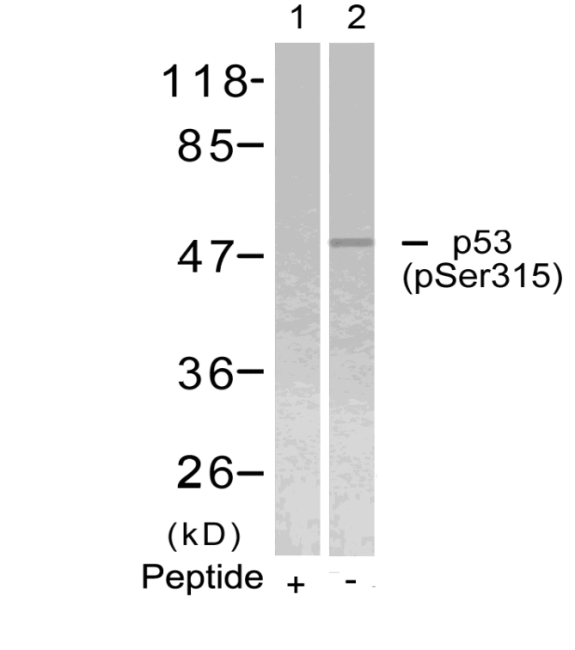 Western blot analysis of extracts from Hela cells using p53 (Phospho-Ser315) Antibody #11100 (Lane 2) and the same antibody preincubated with blocking peptide (Lane1).Validation by various cell lines
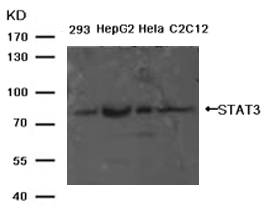
Western blot analysis of extracts from Hela cells using p53 (Phospho-Ser315) Antibody #11100 (Lane 2) and the same antibody preincubated with blocking peptide (Lane1).Validation by various cell lines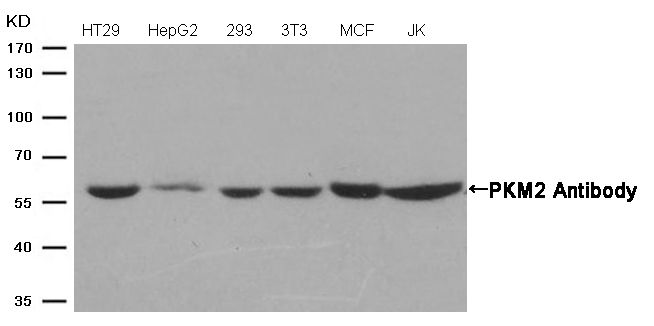 Western blot analysis of extracts from various cells using PKM2 Antibody #21578.
Western blot analysis of extracts from various cells using PKM2 Antibody #21578.

-
New Phospho-specificity Verification with Phosphatase
In order to further verify phospho-specificity and ensure quality, SAB has increased phosphatase treatment method in validation of phospho-specific antibodies. Up to the present SAB has used numerous analytical methods in antibody characterization.
Western blotting(WB)
Phosphatase treatment
Stimulus treatment
Blocking peptide control
Loading control
Validation by various cell lines
Immunohistochemistry(IHC)
Blocking peptide control
Validation with different tissues
Immunofluorescence / Immunocytochemistry(IF/ICC)
Validation by various cell lines
Subcellular localization and co-localization analysis
Flow cytometry(FCM)
ELISA
Akt(Phospho-Ser473) Antibody 11054 Western blot analysis of extracts from 293 cells, treatedwith EGF or calf intestinal phosphatase (CIP), using Akt(Phospho-Ser473) Antibody #11054. MORE> 
PTEN(Phospho-Ser380/Thr382/Thr383) Antibody 11056 Western blot analysis of extracts from Hela cells, treated with IFNa or calf intestinal phosphatase (CIP), using PTEN (Phospho-Ser380/Thr382/Thr383) Antibody #11056. MORE> 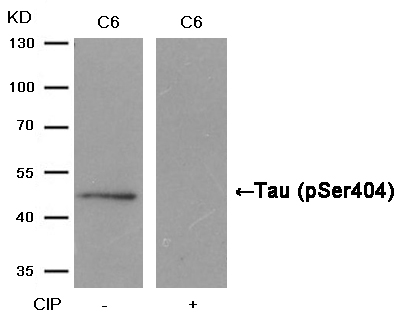
Tau(Phospho-Ser404) Antibody 11112 Western blot analysis of extracts from C6 cells, treated with calf intestinal phosphatase (CIP), using Tau (Phospho-Ser404) Antibody #11112. MORE> 
NFkB-p65(Phospho-Ser529) Antibody 11217 Western blot analysis of extracts from MCF cells, treated with TNFa+CA or calf intestinal phosphatase (CIP), using NFκB-p65 (Phospho-Ser529) Antibody #11217. MORE> 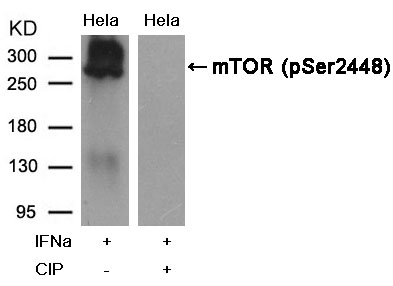
mTOR(Phospho-Ser2448) Antibody 11221 Western blot analysis of extracts from Hela cells, treated with IFNa or calf intestinal phosphatase (CIP), using mTOR (Phospho-Ser2448) Antibody #11221. MORE> 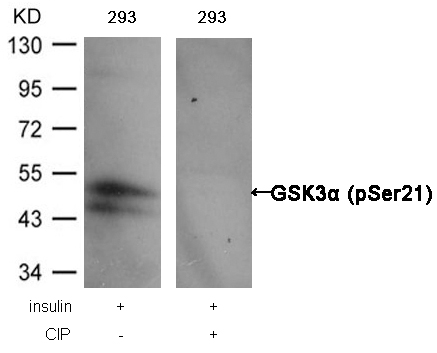
GSK3α (Phospho-Ser21) Antibody 11007 Western blot analysis of extracts from 293 cells, treated with insulin or calf intestinal phosphatase (CIP), using GSK3α (Phospho-Ser21) Antibody #11007. MORE> 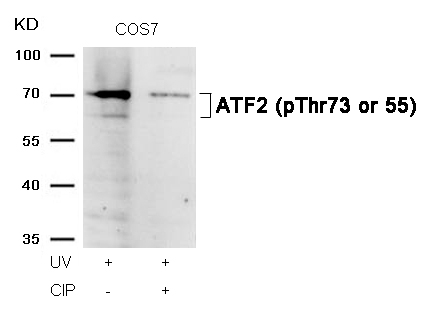
ATF2 (Phospho-Thr73 or 55) Antibody 11032 Western blot analysis of extracts from COS7 cells, treated with UV or calf intestinal phosphatase (CIP), using ATF2 (Phospho-Thr73 or 55) Antibody #11032. MORE> 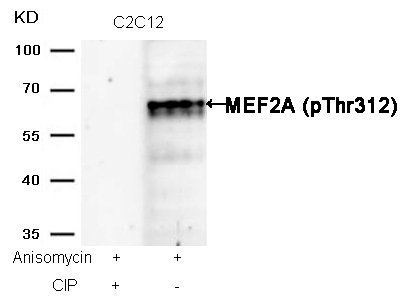
MEF2A (Phospho-Thr312) Antibody 11039 Western blot analysis of extracts from C2C12 cells, treated with Anisomycin or calf intestinal phosphatase (CIP), using MEF2A (Phospho-Thr312) Antibody #11039. MORE> 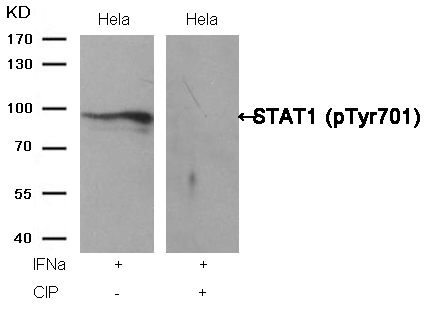
STAT1 (Phospho-Tyr701) Antibody 11044 Western blot analysis of extracts from Hela cells, treated with IFNa or calf intestinal phosphatase (CIP), using STAT1 (Phospho-Tyr701) Antibody #11044. MORE> 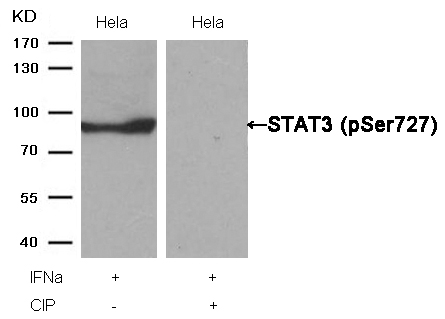
STAT3 (Phospho-Ser727) Antibody 11046 Western blot analysis of extracts from Hela cells, treated with IFNa or calf intestinal phosphatase (CIP), using STAT3 (Phospho-Ser727) Antibody #11046. MORE> 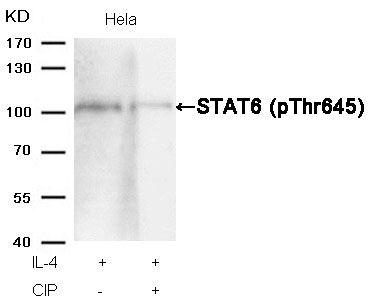
STAT6 (Phospho-Thr645) Antibody 11051 Western blot analysis of extracts from Hela cells, treated with IL-4 or calf intestinal phosphatase (CIP), using STAT6 (Phospho-Thr645) Antibody #11051. MORE> 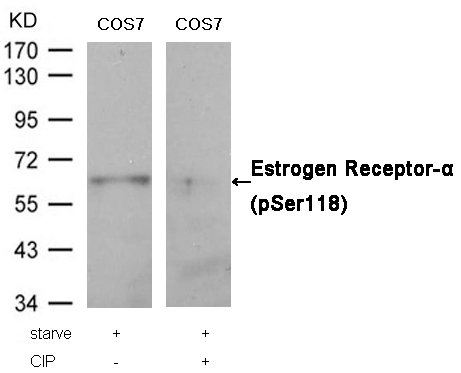
Estrogen Receptor-α (Phospho-Ser118) Antibody 11072 Western blot analysis of extracts from COS7 cells, treated with starve or calf intestinal phosphatase (CIP), using Estrogen Receptor-α (Phospho-Ser118) Antibody #11072. MORE> 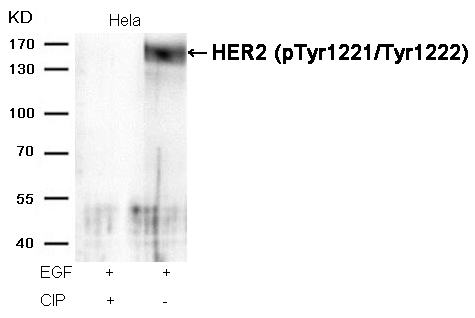
HER2 (Phospho-Tyr1221/Tyr1222) Antibody 11076 Western blot analysis of extracts from Hela cells, treated with EGF or calf intestinal phosphatase (CIP), using HER2 (Phospho-Tyr1221/Tyr1222) Antibody #11076. MORE> 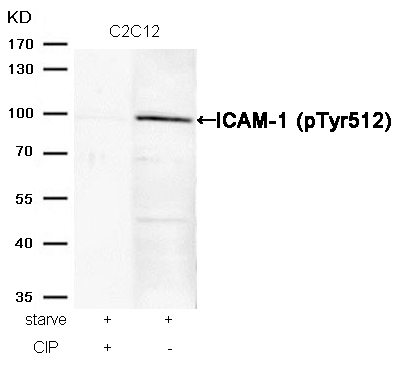
ICAM-1 (Phospho-Tyr512) Antibody 11083 Western blot analysis of extracts from C2C12 cells, treated with starve or calf intestinal phosphatase (CIP), using ICAM-1 (Phospho-Tyr512) Antibody #11083. MORE> 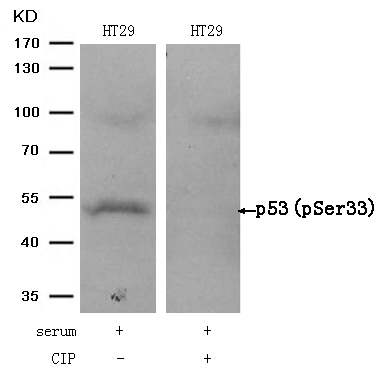
p53 (Phospho-Ser33) Antibody 11097 Western blot analysis of extracts from HT29 cells, treated with serum or calf intestinal phosphatase (CIP), using p53 (Phospho-Ser33) Antibody #11097. MORE> 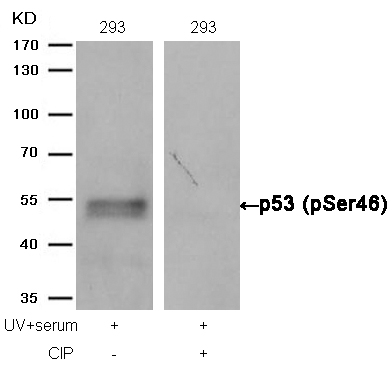
p53 (Phospho-Ser46) Antibody 11099 Western blot analysis of extracts from 293 cells, treated with UV+serum or calf intestinal phosphatase (CIP), using p53 (Phospho-Ser46) Antibody #11099. MORE> 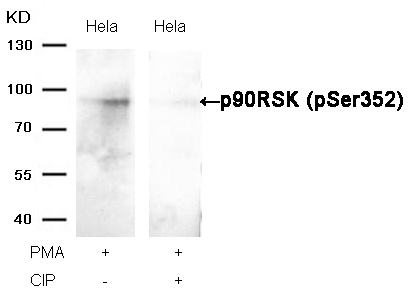
p90RSK (Phospho-Ser352) Antibody 11113 Western blot analysis of extracts from Hela cells, treated with PMA or calf intestinal phosphatase (CIP), using p90RSK (Phospho-Ser352) Antibody #11113. MORE> 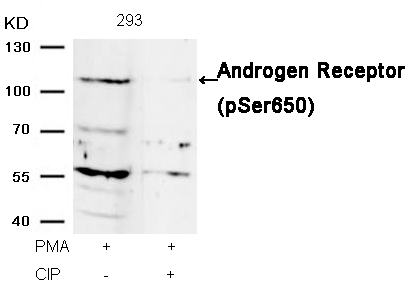
Androgen Receptor (Phospho-Ser650) Antibody 11120 Western blot analysis of extracts from 293 cells, treated with PMA or calf intestinal phosphatase (CIP), using Androgen Receptor (Phospho-Ser650) Antibody #11120. MORE> 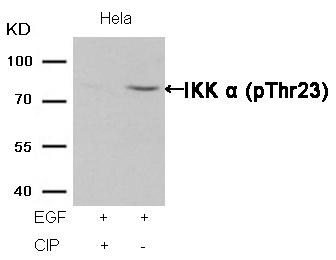
IKK α (Phospho-Thr23) Antibody 11129 Western blot analysis of extracts from Hela cells, treated with EGF or calf intestinal phosphatase (CIP), using IKK α (Phospho-Thr23) Antibody #11129. MORE> 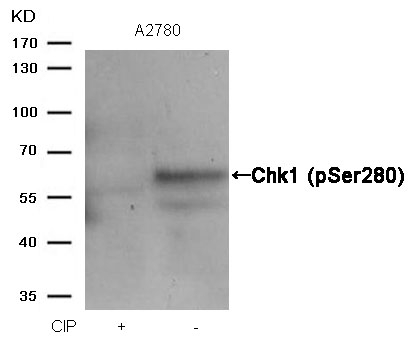
Chk1 (Phospho-Ser280) Antibody 11140 Western blot analysis of extracts from A2780 cells, treated with calf intestinal phosphatase (CIP), using Chk1 (Phospho-Ser280) Antibody #11140. MORE> 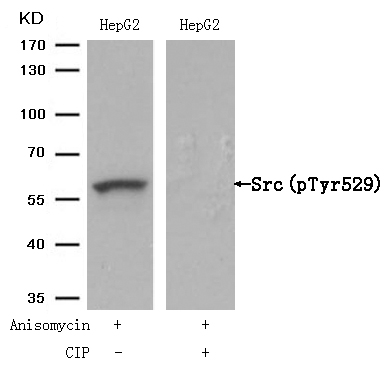
Src(Phospho-Tyr529) Antibody 11153 Western blot analysis of extracts from HepG2 cells, treated with Anisomycin or calf intestinal phosphatase (CIP), using Src (Phospho-Tyr529) Antibody #11153. MORE> 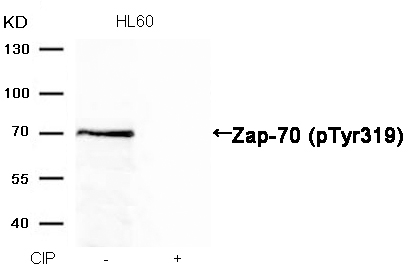
Zap-70 (Phospho-Tyr319) Antibody 11159 Western blot analysis of extracts from HL60 cells, treated with calf intestinal phosphatase (CIP), using Zap-70 (Phospho-Tyr319) Antibody #11159. MORE> 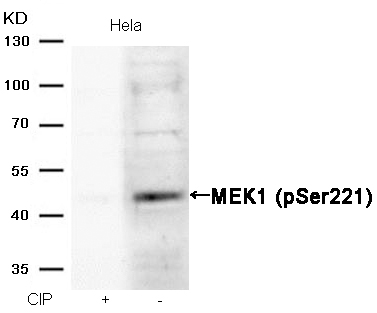
MEK1 (Phospho-Ser221) Antibody 11161 Western blot analysis of extracts from Hela cells, treated with calf intestinal phosphatase (CIP), using MEK1 (Phospho-Ser221) Antibody #11161. MORE> 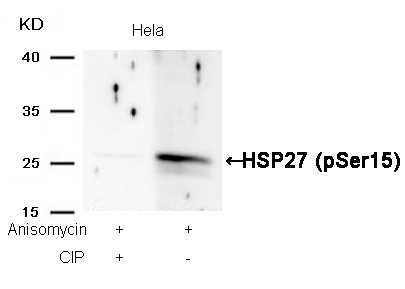
HSP27 (Phospho-Ser15) Antibody 11164 Western blot analysis of extracts from Hela cells, treated with Anisomycin or calf intestinal phosphatase (CIP), using HSP27 (Phospho-Ser15) Antibody #11164. MORE> 
AMPKα1 (Phospho-Ser487)Antibody 11174 Western blot analysis of extracts from C6 cells, treated with EGF or calf intestinal phosphatase (CIP), using AMPKα1 (Phospho-Ser487) Antibody #11174. MORE> 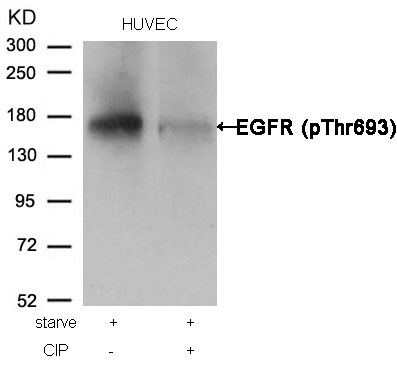
EGFR (Phospho-Thr693) Antibody 11187 Western blot analysis of extracts from HUVEC cells, treated with starve or calf intestinal phosphatase (CIP), using EGFR (Phospho-Thr693) Antibody #11187. MORE> 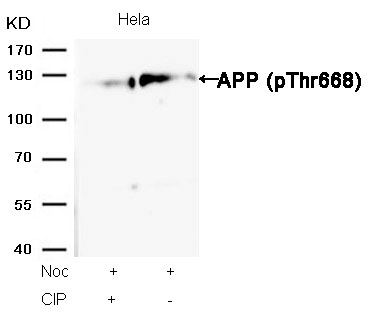
APP (Phospho-Thr668) Antibody 11190 Western blot analysis of extracts from Hela cells, treated with Noc or calf intestinal phosphatase (CIP), using APP (Phospho-Thr668) Antibody #11190. MORE> 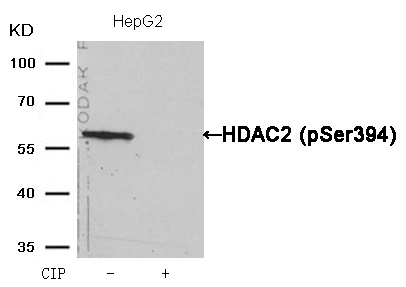
HDAC2 (Phospho-Ser394) Antibody 11191 Western blot analysis of extracts from HepG2 cells, treated with calf intestinal phosphatase (CIP), using HDAC2 (Phospho-Ser394) Antibody #11191. MORE> 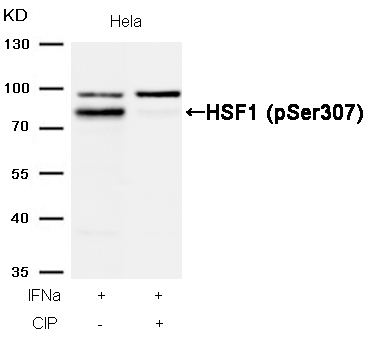
HSF1 (Phospho-Ser307) Antibody 11195 Western blot analysis of extracts from Hela cells, treated with IFNa or calf intestinal phosphatase (CIP), using HSF1 (Phospho-Ser307) Antibody #11195. MORE> 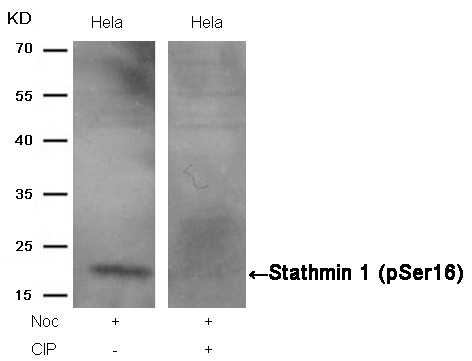
Stathmin 1 (Phospho-Ser16) Antibody 11234 Western blot analysis of extracts from Hela cells, treated with Noc or calf intestinal phosphatase (CIP), using Stathmin 1 (Phospho-Ser16) Antibody #11234. MORE> 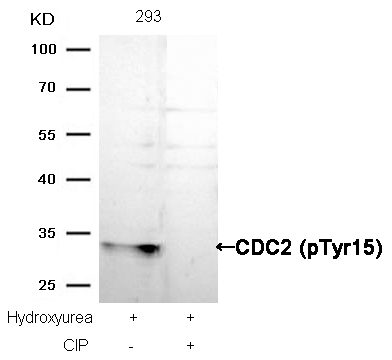
CDC2 (Phospho-Tyr15) Antibody 11244 Western blot analysis of extracts from Hela cells, treated with Noc or calf intestinal phosphatase (CIP), using Stathmin 1 (Phospho-Ser16) Antibody #11244. MORE> 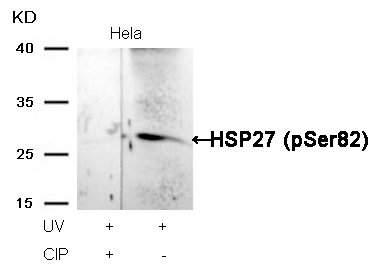
HSP27 (Phospho-Ser82) Antibody 11248 Western blot analysis of extracts from Hela cells, treated with UV or calf intestinal phosphatase (CIP), using HSP27 (Phospho-Ser82) Antibody #11248. MORE> 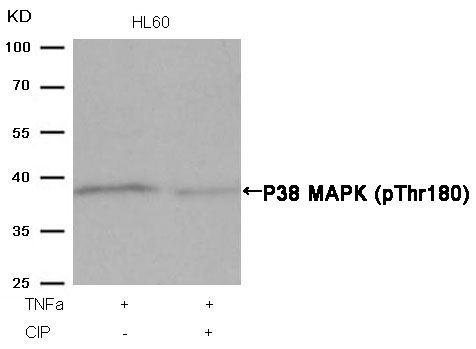
P38 MAPK (Phospho-Thr180) Antibody 11252 Western blot analysis of extracts from HL60 cells, treated with TNFa or calf intestinal phosphatase (CIP), using P38 MAPK (Phospho-Thr180) Antibody #11252. MORE> 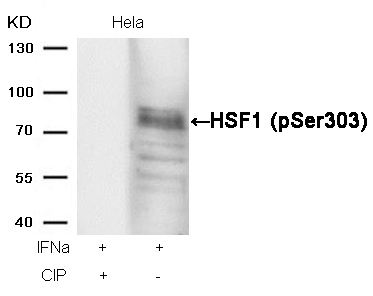
HSF1 (phospho-Ser303) Antibody 11263 Western blot analysis of extracts from Hela cells, treated with IFNa or calf intestinal phosphatase (CIP), using HSF1 (phospho-Ser303) Antibody #11263. MORE> 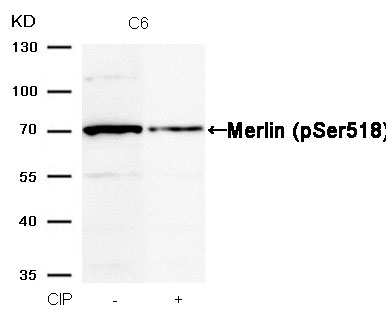
Merlin (Phospho-Ser518) Antibody 11266 Western blot analysis of extracts from C6 cells, treated with calf intestinal phosphatase (CIP), using Merlin (Phospho-Ser518) Antibody #11266. MORE> 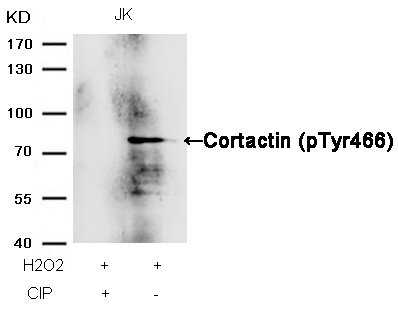
Cortactin (Phospho-Tyr466) Antibody 11272 Western blot analysis of extracts from JK cells, treated with H2O2 or calf intestinal phosphatase (CIP), using Cortactin (Phospho-Tyr466) Antibody #11272. MORE> 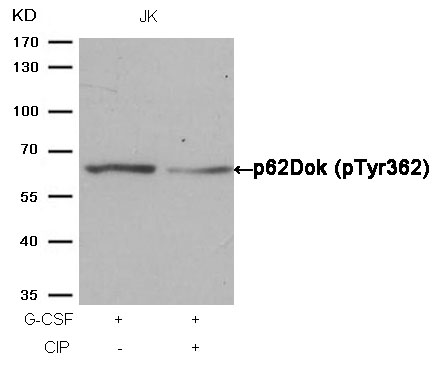
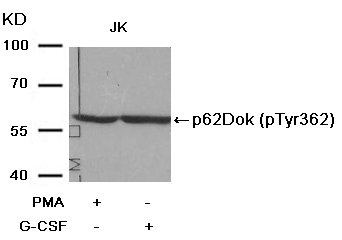
p62Dok (phospho-Tyr362) Antibody 11276 Western blot analysis of extracts from JK cells, treated with G-CSF or calf intestinal phosphatase (CIP), using p62Dok (phospho-Tyr362) Antibody #11276. MORE> 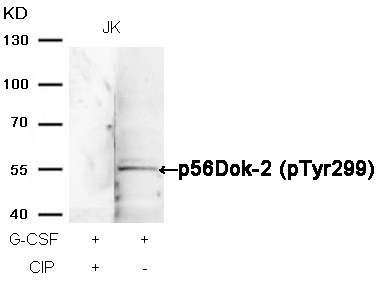
p56Dok-2 (Phospho-Tyr299) Antibody 11278 Western blot analysis of extracts from JK cells, treated with G-CSF or calf intestinal phosphatase (CIP), using p56Dok-2 (Phospho-Tyr299) Antibody #11278. MORE> 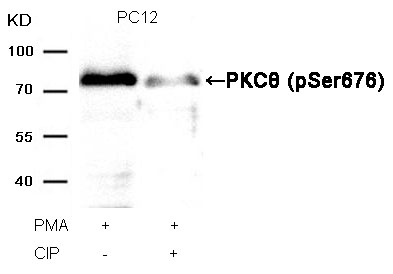
PKCθ (Phospho-Ser676) Antibody 11297 Western blot analysis of extracts from PC12 cells, treated with PMA or calf intestinal phosphatase (CIP), using PKCθ (Phospho-Ser676) Antibody #11297. MORE> 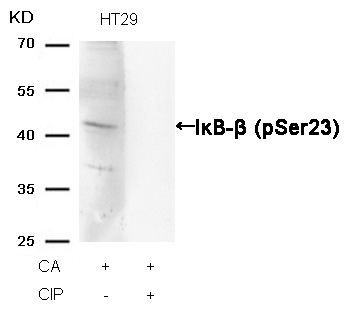
IκB-β (Phospho-Ser23) Antibody 11304 Western blot analysis of extracts from HT29 cells, treated with CA or calf intestinal phosphatase (CIP), using IκB-β (Phospho-Ser23) Antibody #11304. MORE> 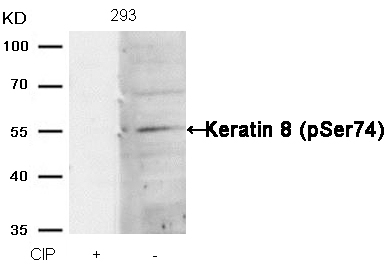
Keratin 8 (Phospho-Ser74) Antibody 11307 Western blot analysis of extracts from 293 cells, treated with calf intestinal phosphatase (CIP), using Keratin 8 (Phospho-Ser74) Antibody #11307. MORE> 
Myc (Phospho-Ser62) Antibody 11311 Western blot analysis of extracts from K562 cells, treated with calf intestinal phosphatase (CIP), using Myc (Phospho-Ser62) Antibody #11311. MORE> 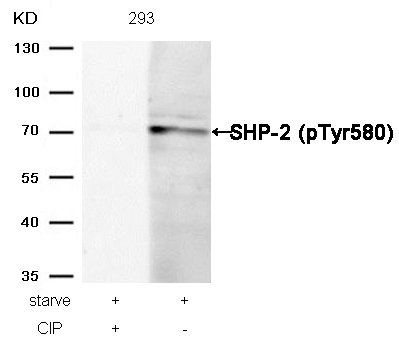
SHP-2 (Phospho-Tyr580) Antibody 11320 Western blot analysis of extracts from 293 cells, treated with starve or calf intestinal phosphatase (CIP), using SHP-2 (Phospho-Tyr580) Antibody #11320. MORE> 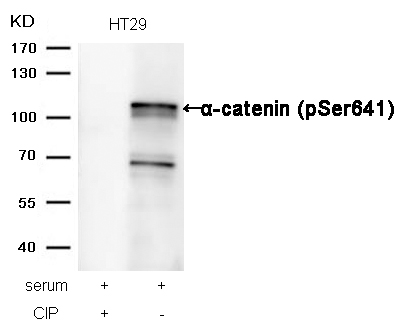
α-catenin (Phospho-Ser641) Antibody 11330 Western blot analysis of extracts from HT29 cells, treated with serum or calf intestinal phosphatase (CIP), using α-catenin (Phospho-Ser641) Antibody #11330. MORE> 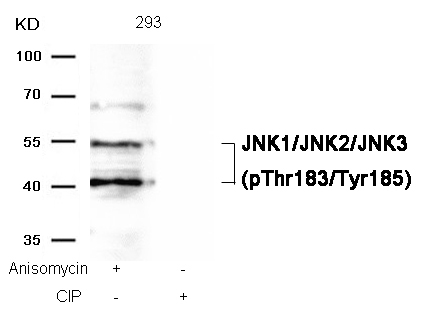
JNK1/JNK2/JNK3 (phospho-Thr183/Tyr185) Antibody 11504 Western blot analysis of extracts from 293 cells, treated with Anisomycin or calf intestinal phosphatase (CIP), using JNK1/JNK2/JNK3 (phospho-Thr183/Tyr185) Antibody #11504. MORE> 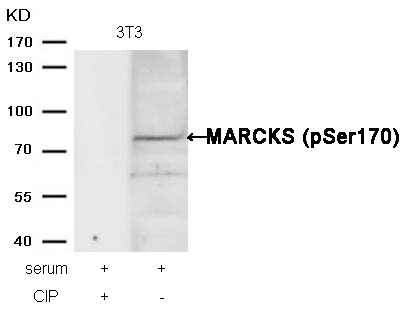
MARCKS (phospho-Ser170) Antibody 11535 Western blot analysis of extracts from 3T3 cells, treated with serum or calf intestinal phosphatase (CIP), using MARCKS (phospho-Ser170) Antibody #11535. MORE> 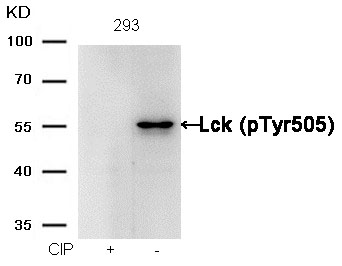
Lck (phospho-Tyr505) Antibody 11537 Western blot analysis of extracts from 293 cells, treated with calf intestinal phosphatase (CIP), using Lck (phospho-Tyr505) Antibody #11537. MORE> 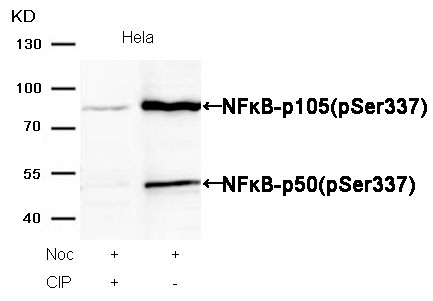
NFκB-p105/p50(Phospho-Ser337) Antibody 11017 Western blot analysis of extracts from Hela cells, treated with Noc or calf intestinal phosphatase (CIP), using NFκB-p105/p50(Phospho-Ser337) Antibody #11017. MORE> -
New Published Paper
Volume 35, Issue 1, January 2014, Pages 316–326The use of hollow mesoporous silica nanospheres to encapsulate bortezomib and improve efficacy for non-small cell lung cancer therapyJia Shena, b, c, 1, Guosheng Songd, 1, Man Ana, c, Xianqian Lie, Ning Wue, Kangcheng Ruana, c, Junqing Hud, Ronggui Hua, c,a State Key Laboratory of Molecular Biology, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, 320 Yue-yang Road, Shanghai 200031, Chinab University of Chinese Academy of Sciences, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, 320 Yue-yang Road, Shanghai 200031, Chinac Shanghai Key Laboratory of Molecular Andrology, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, 320 Yue-yang Road, Shanghai 200031, Chinad State Key Laboratory for Modification of Chemical Fibers and Polymer Materials, College of Materials Science and Engineering, Donghua University, Shanghai 201620, Chinae Department of Clinical Oncology, Xuhui Central Hospital, 966 Middle Huaihai Road, Shanghai 200031, ChinaAbstractBortezomib (BTZ) is the first clinically approved proteasome inhibitor for treating multiple human malignancies. However, the poor water-solubility and low stability of BTZ and the emergence of tumor resistance have severely restrained its therapeutic efficacy. Herein, we report the application of hollow mesoporous silica nanospheres (HMSNs) in encapsulating BTZ for drug delivery. In in vitro cell viability assay on human NSCLC H1299 cells, the half-maximum inhibiting concentration (IC50) of HMSNs–BTZ was 42% of that for free BTZ in 48 h treatments. In vivo tumor-suppression assay further indicated that HMSNs–BTZ (0.3 mg/kg) showed approximately 1.5 folds stronger anti-tumor activity than free BTZ. Furthermore, we report that more potent induction of cell cycle arrest and apoptotic cell death, along with promoted activation of Caspase 3 and autophagy might mechanistically underlie the improved anti-tumor efficacy of HMSNs–BTZ. Finally, the tumor-suppressing effect of HMSNs–BTZ was enhanced in the presence of wild-type p53 signaling, suggesting a potential enhancement in clinical efficacy with combined p53 gene therapy and BTZ-based chemotherapy. Therefore, the HMSNs-based nanoparticles are emerging as a promising platform to deliver therapeutic agents for beneficial clinical outcomes through lowering doses and frequency of drug administration and reducing potential side effects.KeywordsAnti-tumor activity; Apoptotic cell death; Bortezomib; Drug delivery; Mesoporous silica; NSCLC





